PartSeg is gui and library for segmentation algorithms.
This application is designed to help biologist with segmentation based on threshold and connected components
- Tutorial: Chromosome 1 (as gui) link
- Data for chromosome 1 tutorial link
- Tutorial: Different neuron types (as library) link
- From binaries:
- With pip (on linux you need to install
numpyandcythonearlier)- From pypi:
pip install PartSeg - From repository:
git clone [email protected]:4DNucleome/PartSeg.gitcd PartSeg/pip install -e .
- From pypi:
If you download binaries look for PartSeg_exec file inside the PartSeg folder
If you install from repository or from pip you cat run it with PartSeg command or python -m PartSeg.
First option do not work on Windows.
PartSeg export few commandline options:
--no_report- disable reporting errors to authors--no_dialog- disable reporting errors to authors and showing error dialog. Use only when running from terminal.segmentation_analysis- skip launcher and start analysis guisegmentation- skip launcher and start segmentation gui
Current version of PartSeg use tifffile package to read *.tiff files. Because newer version is easy to install
only on Windows. If you install it manually with imagecodecs it should work.
Saved project are tar files compressed with gzip or bz2
Metadata are saved in data.json file (in json format) images/mask are saved as *.npy (numpy array format)
Launcher. Chose program that you will launch:
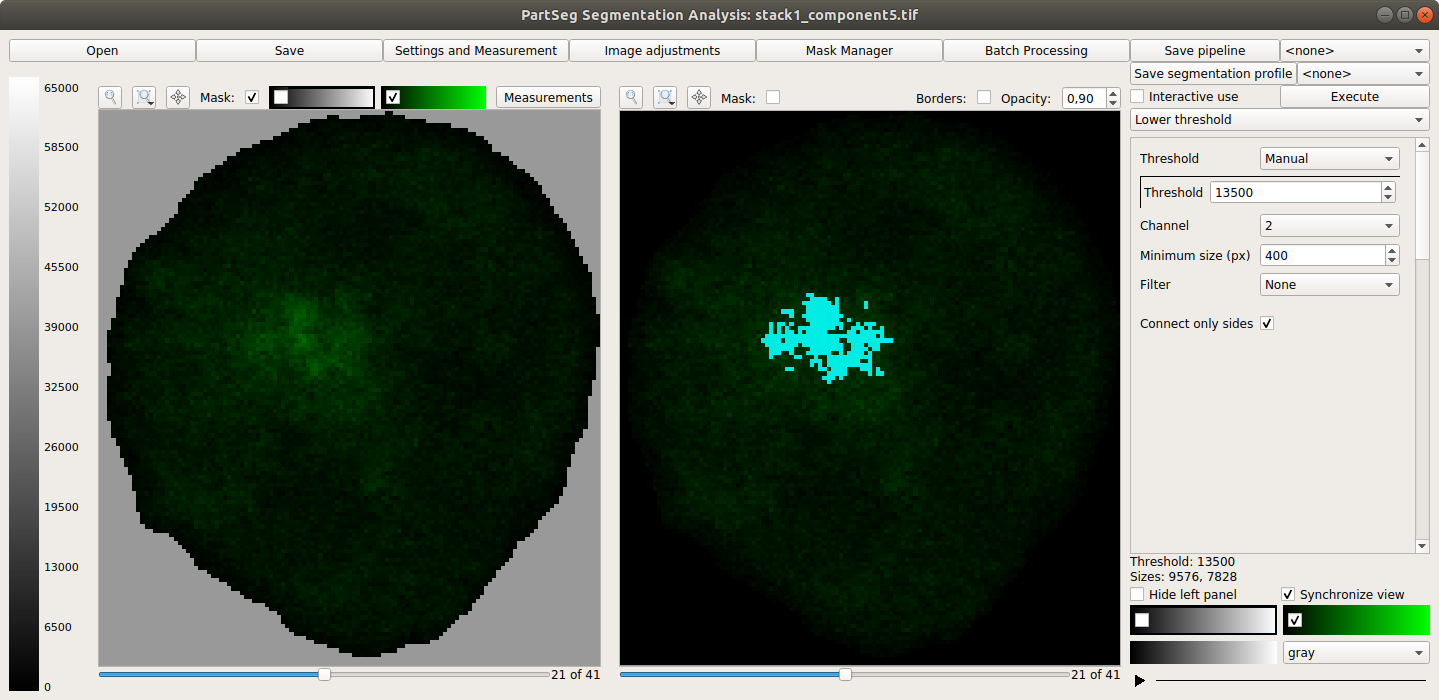
Main window of Segmentation Analysis:
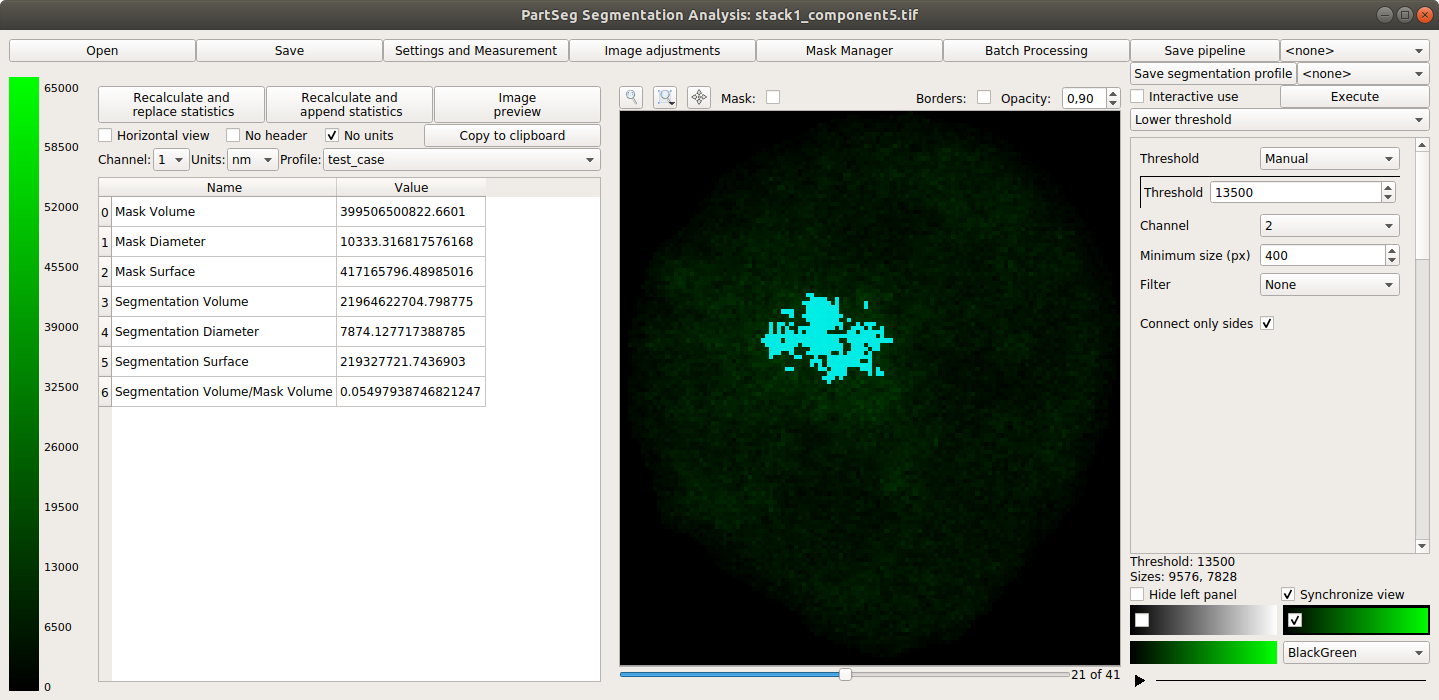
Main window of Segmentation Analysis with view on measurement result:
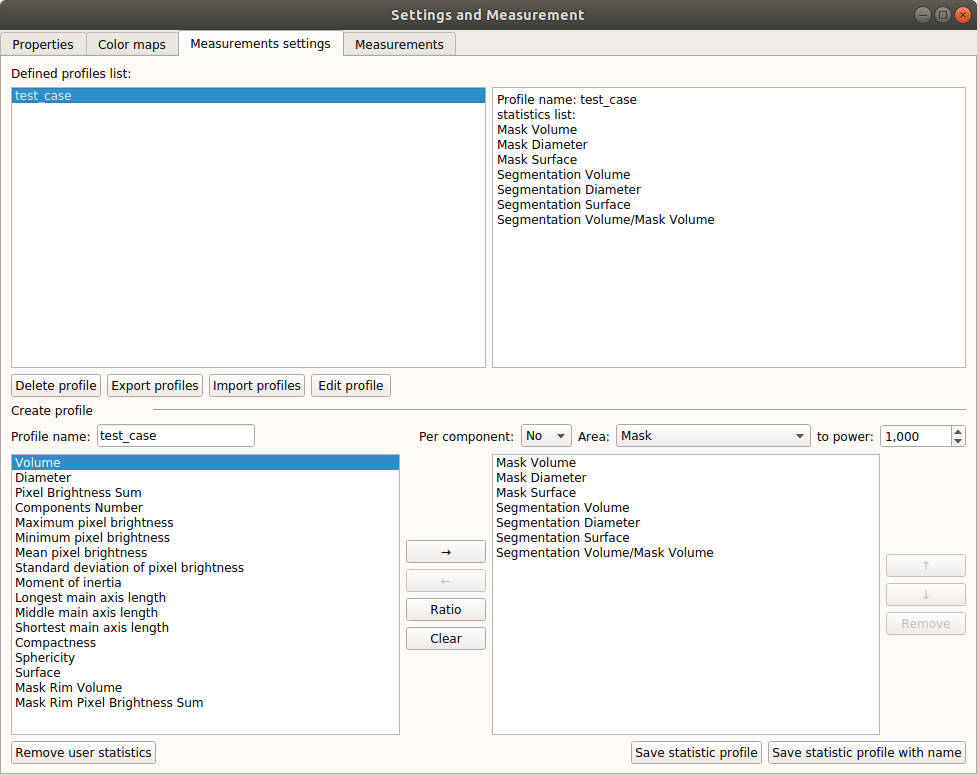
Window for creating set of measurements:
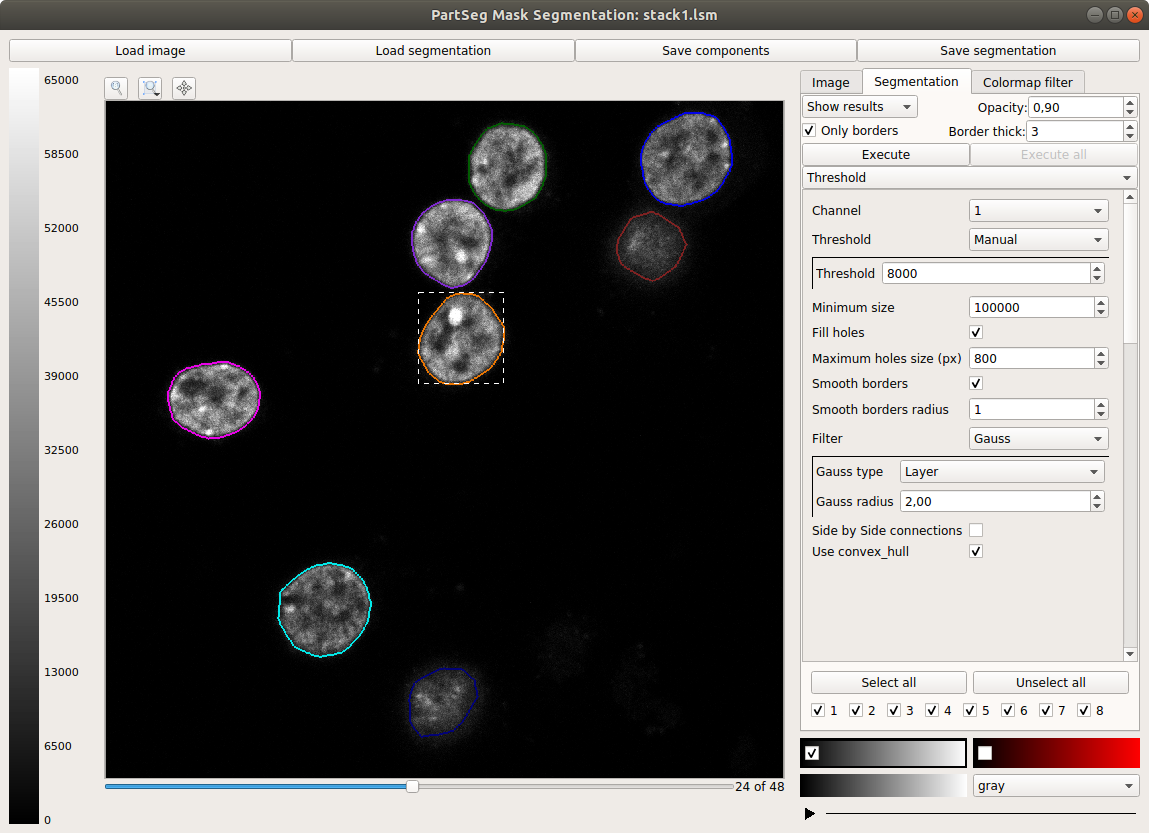
Main window of Mask Segmentation:
Laboratory of functional and structural genomics http://4dnucleome.cent.uw.edu.pl/