The following example uses data from the Consortium for Neuropsychiatric Phenomics (CNP) dataset described in (Poldrack et al. 2016). This data was obtained from the OpenfMRI database. Its accession number is ds000030. In this example we work from the preprocessed data release v1.0.4. The release includes both structural outputs preprocessed with freesurfer (version 6.0.0, (Fischl 2012)) and resting state fMRI data preprocessed with the fmriprep pipeline (see (Gorgolewski et al. 2017)).
Where /path/to/mount/data/to is a local folder on your machine
docker run -it -p 8888:8888 --rm -v /path/to/mount/data/to:/home/data mmanogaran/cifti
fy-demo:0.1Example (from my Windows machine):
docker run -it -p 8888:8888 --rm -v C:\Users\edickie\ciftify_demo_test\:/home/data mmanogaran/cifti
fy-demo:0.1We will download and unzip the freesurfer and fmriprep outputs from subjects 50004-50008
mkdir -p /home/data/src_data
cd /home/data/src_data ## change this to the location of the files on your system
## the fmriprep outputs
wget https://s3.amazonaws.com/openneuro/ds000030/ds000030_R1.0.4/compressed/ds000030_R1.0.4_derivatives_sub50004-50008.zip
unzip ds000030_R1.0.4_derivatives_sub50004-50008.zip
wget https://s3.amazonaws.com/openneuro/ds000030/ds000030_R1.0.4/compressed/ds000030_R1.0.4_derivatives_freesurfer_sub50004-50008.zip
unzip ds000030_R1.0.4_derivatives_freesurfer_sub50004-50008.zipThis outputs are organized into BIDS data structure for "derivatives" calculated in this manner. Speciftically, all outputs are nested within subfolders "freesurfer" (for freesurfer) and "fmriprep" (for fmri), which are sitting within ds000030_R1.0.4/derivatives. Within the freesurfer subfolder is the full freesurfer recon-all data structure - where subject id is the top level and folders (label, mri, etc) sit below that. The fmriprep derivatives also are organized into a subfolder for each participant. Within these folders sit anatomical ("anat") and functional ("func") derivatives. For ciftify, we require the preprocessed fmri data in T1w ("native") space. In this example, we will select the resting state run. For participant sub-50005, this file is named "sub-50005_task-rest_bold_space-T1w_preproc.nii.gz"
ds000030_R1.0.4
└── derivatives
├── fmriprep
│ ├── sub-50005
| │ ├── anat
│ │ └── func
| | ├── sub-50004_task-rest_bold_space-T1w_preproc.nii.gz
| | └── ...
│ ├── sub-50006...
│ └── sub-...
└── freesurfer
├── sub-50005
│ ├── label
│ ├── mri
│ ├── scripts
│ ├── stats
│ ├── surf
│ ├── tmp
│ ├── touch
│ └── trash
├── sub-50006...
└── sub-...
For installation instructions visit the installation page.
If you are in a tutorial there is probably a local install of the ciftify package to you learn with. Visit this page for instructions to source the environment.
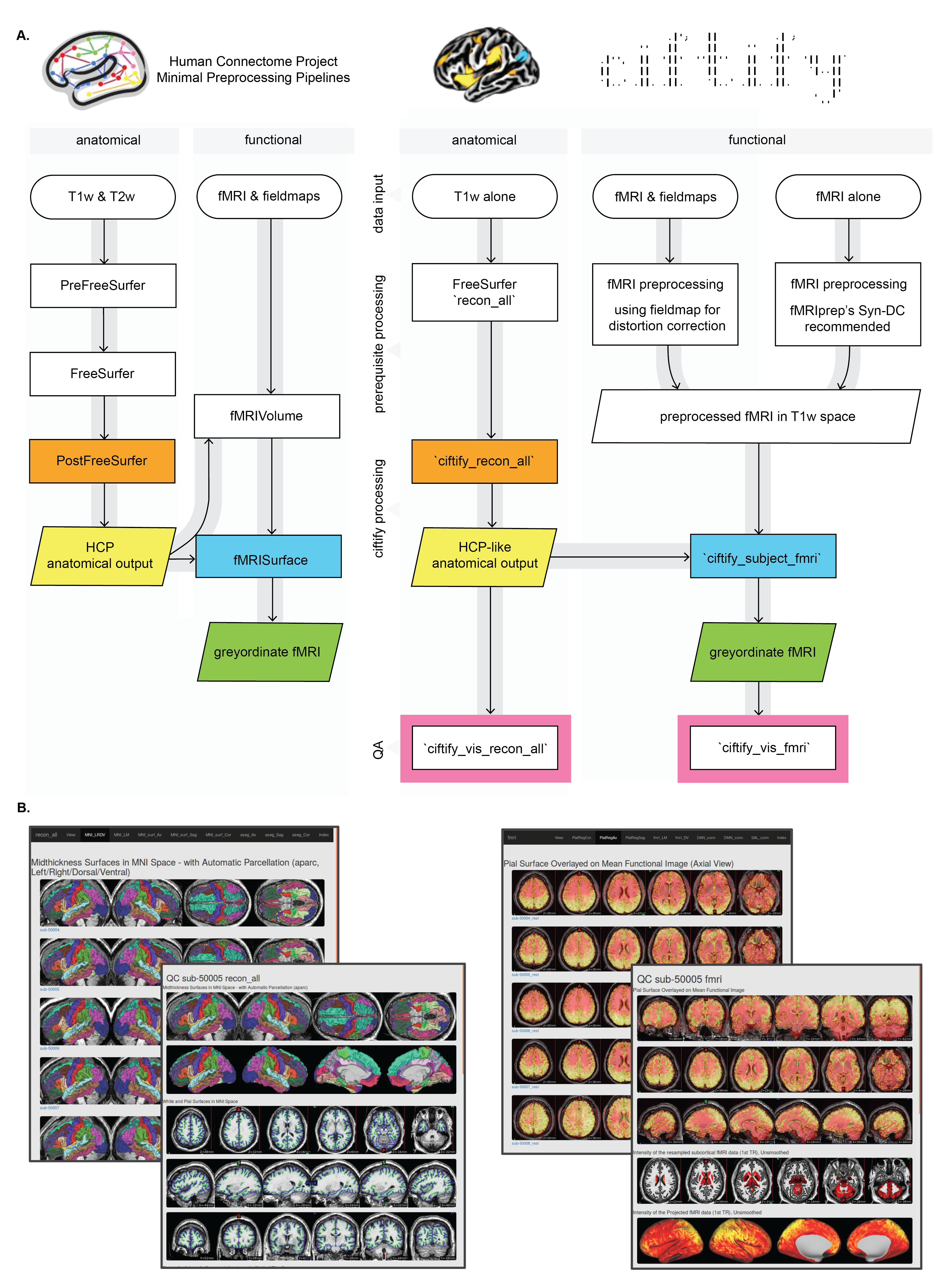
This step converts a subjects freesurfer output into an HCP-like structural anatomy output.
ciftify_recon_all --ciftify-work-dir /home/data/ciftify_demo_01 --fs-subjects-dir /home/data/src_data/ds000030_R1.0.4/derivatives/freesurfer sub-50005Note: this steps takes hours (now that the default behaviour is MSMSulc). See the
ciftify_demo_02 folder for the expected outputs. We are going to continue from this stage.
After we convert the files we should check the quality of the outputs. The following functions will create snapshots for quality assurance and html pages for visualizing them together
Note: the subject step is run once per participant. The index step can be run once at the end.
cifti_vis_recon_all subject --ciftify-work-dir /home/data/ciftify_demo_02 sub-50005
cifti_vis_recon_all index --ciftify-work-dir /home/data/ciftify_demo_02Now that we have the surfaces, we can use ciftify_subject_fmri to map our preprocessed fMRI data to our subjects' surfaces (as well as resample the subcortical data).
Note: the last argument should be unique to this specific scan. Therefore under BIDS, if appropriate, this name should also contain session, run, and any other relevant naming. (i.e. ses-01_task-rest_run-01)
ciftify_subject_fmri --ciftify-work-dir /home/data/ciftify_demo_02 /home/data/src_data/ds000030_R1.0.4/derivatives/fmriprep/sub-50005/func/sub-50005_task-rest_bold_space-T1w_preproc.nii.gz sub-50005 ses-01_task-rest_run-01Note: this steps approx 12 minutes to run. See the
ciftify_demo_03 folder for the expected outputs. We are going to continue from this stage.
The next steps generates quality assurance images for the fmri data.
cifti_vis_fmri subject --ciftify-work-dir /home/data/ciftify_demo_03 ses-01_task-rest_run-01 sub-50005
cifti_vis_fmri index --ciftify-work-dir /home/data/ciftify_demo_03A zip file of the expected outputs for sub-50005 as well as QA images for sub-50004-sub-50008 can be downloaded from here.
ciftify_PINT_vertices [options] <func.dtseries.nii> <left-surface.gii> <right-surface.gii> <input-vertices.csv> <outputprefix>
ciftify_PINT_vertices [options] <func.dtseries.nii> <left-surface.gii> <right-surface.gii> <input-vertices.csv> <outputprefix>
Use wb_command to define a left putamen roi from the participants' freesurfer automatic segmentation
cd /home/data/ciftify_demo_03/sub-50005/MNINonLinear
wb_command -volume-label-to-roi \
wmparc.nii.gz \
right_putamen_roi.nii.gz \
-name "RIGHT-PUTAMEN"Run a seed correlation with this mask and the cifti functional file
ciftify_seed_corr \
Results/ses-01_task-rest_run-01/ses-01_task-rest_run-01_Atlas_s0.dtseries.nii \
right_putamen_roi.nii.gzGenerate a report from this seed correlation map --This is still a WIP...
cd Results/ses-01_task-rest_run-01
ciftify_statclust_report \
--min-threshold -0.5 \
--max-threshold 0.5 \
--output-peaks \
ses-01_task-rest_run-01_Atlas_s0_right_putamen_roi.dscalar.nii