Effectively this are (almost [1] [2]) all capabilities of the SCOPE model.
This is an input structure that controls the workflow.
The values have binary (or tertiary) logic thus equal to 0 or 1 (or 2).
Influence on the output files is highlighted in the corresponding section :ref:`outfiles:Output files`
Note
Not all combinations can bring to the desired result
SCOPE.m: read from setoptions.csv
Defines rules of input reading
Switch in SCOPE.m (multiple)
- 0
- individual run(s): specify one value for fixed input parameters, and an equal number (> 1) of values for all parameters that vary between the runs.
- 1
- time series (uses text files with meteo input as time series from "./input/dataset X" with files similar to ./input/:ref:`directories/input:dataset for_verification` specified in the
filenames.csv - 2
- Lookup-Table: specify a number of values in the row of input parameters. All possible combinations of inputs will be used.
Let us illustrate what the difference is in details.
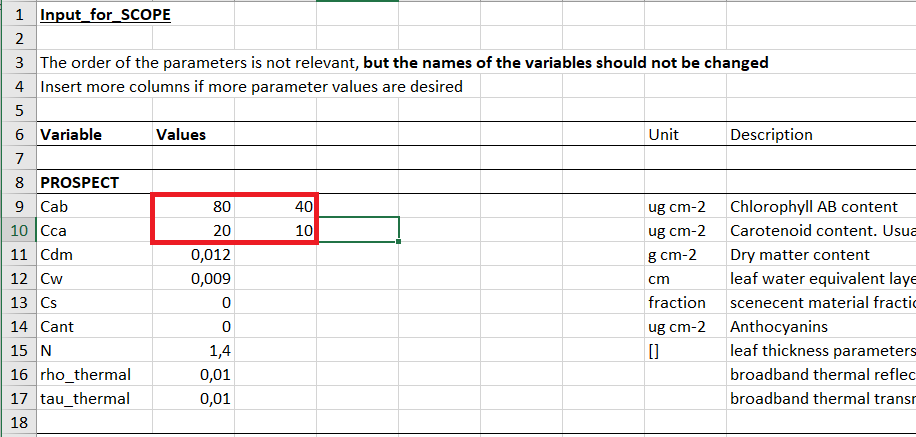
It is possible to specify several values in a row on input_data.csv. Suppose we have an the following combination of input parameters. Notice, we provide two values for Cab and Cca parameters.
If individual run(s) (options.simulation == 0) was chosen the given combination will end up in two simulations:
- Cab=80, Cca=20
- Cab=40, Cca=10
If Lookup-Table (options.simulation == 2) was chosen the given combination will end up in four simulations:
- Cab=80, Cca=20
- Cab=80, Cca=10
- Cab=40, Cca=20
- Cab=40, Cca=10
Calculate soil reflectance or use from a file in ./input/:ref:`directories/input:soil_spectra`
Switch in SCOPE.m
0
use soil spectrum from the file with :ref:`structs/input/soil:soil`.spectrumdefault file issoilnew.txt, can be changed on thefilenamessheetsoil_filecellvariable name isrsfile
- 1
- simulate soil spectrum with the BSM model (:func:`BSM`) parameters are fixed in code
Method of ground heat flux (G) calculation. In soil_heat_method 0 and 1 soil thermal inertia (GAM) is calculated from inputs.
Switch in SCOPE.m, :func:`.select_input`, :func:`.ebal`
0
standard calculation of thermal inertia from soil characteristic
- 1
- empirically calibrated formula from soil moisture content :func:`.Soil_Inertia1` in :func:`.select_input`
- 2
- as constant fraction (0.35) of soil net radiation
soil resistance for evaporation from the pore space (rss) and soil boundary layer resistance (rbs)
Switch in :func:`.select_input`
0
use resistance rss and rbs as provided in inputdata :ref:`structs/input/soil:soil`
- 1
- calculate rss from soil moisture content and correct rbs for LAI :func:`.calc_rssrbs`
Switch in SCOPE.m
0
traditional single layer SCOPE
- 1
- multilayer :ref:`mSCOPE:mSCOPE`
Switch in :func:`.RTMo`
0
Normal SCOPE execution with [13 x 36 x nlayers] sunlit leaves
1
Lite SCOPE execution with [nlayers x 1] sunlit leaves, sunlit leaf inclinations are not accounted for
Calculate spectrum of thermal radiation with spectral emissivity instead of broadband
Warning
only effective with calc_ebal == 1
Switch in SCOPE.m, :func:`.calc_brdf`
0
:func:`.RTMt_sb` - broadband brightness temperature is calculated in accordance to Stefan-Boltzman’s equation.
- 1
- Calculation is done per each wavelength thus takes more time than Stefan-Boltzman.
Calculate BRDF and directional temperature for many angles specified in the file: :ref:`directories/input:directional`.
Warning
- only effective with
calc_ebal == 1 - Be patient, this takes some time
Switch in SCOPE.m, :func:`.calc_brdf`
0
- 1
- struct :ref:`structs/output/directional:directional` is loaded from the file :ref:`directories/input:directional`:func:`.calc_brdf` is launched in
SCOPE.m
Calculate dynamic xanthopyll absorption (zeaxanthin) for simulating PRI (photochemical reflectance index)
Warning
- only effective with
calc_ebal == 1
Switch in SCOPE.m
0
- 1
- :func:`.RTMz` is launched in
SCOPE.mand :func:`.calc_brdf` (ifcalc_directional)
Calculation of vertical profiles (per 60 canopy layers).
Corresponding structure :ref:`structs/output/profiles:profiles`
Switch in SCOPE.m, :func:`.RTMo` and :func:`.ebal`
0
Profiles are not calculated
- 1
- Photosynthetically active radiation (PAR) per layer is calculated in :func:`.RTMo`Energy, temperature and photosynthesis fluxes per layer are calculated in :func:`.ebal`Fluorescence fluxes are calculated in :func:`.RTMf` if (
calc_fluor)
Calculation of fluorescence
Switch in SCOPE.m, :func:`.calc_brdf`
0
No fluorescence
- 1
- total emitted fluorescence is calculated by
SCOPE.m
Fluorescence model
Switch in :func:`.ebal`
- 0
- empirical, with sigmoid for Kn: :func:`.biochemical` (Berry-Van der Tol)
- 1
- :func:`.biochemical_MD12` (von Caemmerer-Magnani)
correct Vcmax and rate constants for temperature
Warning
only effective with Fluorescence_model == 0 i.e. for :func:`.biochemical`
Switch in :func:`.ebal`
- 0
- 1
- correction in accordance to Q10 rule
Switch in :func:`.ebal`
- 0
- do not apply Monin-Obukhov atmospheric stability correction
- 1
- apply Monin-Obukhov atmospheric stability correction
verify the results (compare to saved 'standard' output) to test the code for the first time
Switch in SCOPE.m
- 0
- 1
- runs :func:`.output_verification`
Switch in SCOPE.m, :func:`.bin_to_csv`
- 0
- leave .bin files in output folder
- 1
- convert .bin files to .csv with :func:`.bin_to_csv`, delete .bin files
Save files with full spectrum. May reach huge sizes in long time-series.
Switch in :func:`.create_output_files_binary`
- 0
- do not save
- 1
- save
| [1] | extra output variables that are not saved to files (see :ref:`structs:Structs`) are available in the workspace after the model run. |
| [2] | model can be varied by user, please, consult :ref:`api:API` to learn signatures of functions |