The code repository for the following paper:
Paper: Di Jin, Ryan Rossi, Sungchul Kim and Danai Koutra. On Generalizing Static Node Embedding to Dynamic Settings. The Fifteenth International Conference on Web Search and Data Mining (WSDM), Phoenix, AZ, USA, Feb. 2022.
This repository contains an example to adopt MultiLens as the static embedding approach to obtain the dynamic embeddings.
MultiLens takes two files as input, the graph file and the category file. The input files are placed under small_graphs_lp_time/ directory.
The input graph file can be either static or temporal edge list in the following format separated by tab:
<src> <dst> <weight> <timestamp> (optional)
The edge list is assumed to be re-ordered consecutively from 0, i.e., the minimum node ID is 0, and the maximum node ID is <#node - 1>. Some preprocess files are placed under the preprocess/ directory.
The category file is a mapping between the node ID and its type (e.g., IP, cookie, web agent) with the following format separated by tab:
<category> <id_initial> <id_ending>
To run MultiLens correctly, each subgraph file should have its corresponding category file. But this is not required for most other methods such as node2vec, struc2vec, etc.
To run the framework based on the predefined static embedding approach, an example is given in main.sh, and the command is
./main.sh
which will run
main_split_heuristics.py [temp_model] [graph_series] [static_method] [initial_snapshot_id] [agg_factor] [train_num] [test_num] [time_scale] [iteration_id]
The complete list of argumments of this script are described as follows.
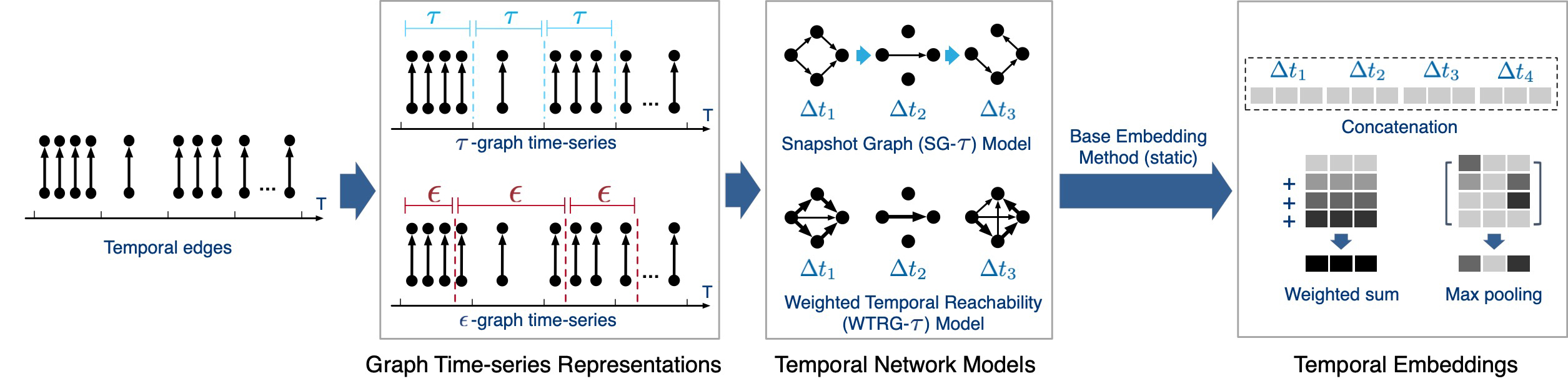
- [temp_model] denotes the specific temporal network model. It supports
<snapshot>and<trg>. To employ the TSG model, one can run our preprocessing code to get one aggregated summary graph and then use the selected embedding approach. - [graph_series] denotes the graph time-series. It supports
<TS>( τ-graph) and<NS>(ε-graph). - [static_method] is the name of static embedding approach. Our code supports the following methods, the original code repo should be placed under
eval/directory. The example code of MultiLens is placed there.struc2vec/graphwave/node2vec/line/struc2vec/multilens/role2vec/graphwave/g2g - [initial_snapshot_id] is the specific snapshot id in the graph time-series to start getting the embeddings.
- [agg_factor] denotes the number of snapshots to aggregate. This is often used with [time_scale].
- [train_num] denote the number of snapshots used to generate the dynamic node embeddings for the link prediction task. In our paper, we set [train_num] as
6to get the dynamic node embeddings and predict links in the next snapshot. - [test_num] denotes the number of snapshots used for testing in the link prediction task.
- [time_scale] denotes the time scale that corresponds to the τ-graph. It supports
day/month. It is often coupled with [agg_factor] to denote other scales. For example, when denoting "week", [time_scale] is setdayand [agg_factor] is set7. - [iteration_id] denotes the specific run id. We conducted 3 runs for all experiments in our paper.
The output dynamic embedding file is generated under the src/emb<iteration_id>/ directory, where <iteration_id> denotes the specific run of experiments. The embedding file follows the general format:
<node_id> [<embedding_values>]
If you encounter any problems running the code, pls feel free to contact Di Jin ([email protected])