The tools of the Human Connectome Project (HCP) adapted for working with non-HCP datasets
ciftify is a set of three types of command line tools:
- conversion tools : bash scripts adapted from HCP Minimal processing pipeline to put preprocessed T1 and fMRI data into an HCP like folder structure
- ciftify tools : Command line tools for making working with cifty format a little easier
- cifti_vis tools : Visualization tools, these use connectome-workbench tools to create pngs of standard views the present theme together in fRML pages.
https://github.com/edickie/ciftify/wiki
First, install the python package and all of its bundled data and scripts. You can do this with a single command with either pip or conda if you have one of them installed. If you don't want to use either of these tools, skip to the 'manual install' step.
To install with pip, type the following in a terminal.
pip install https://github.com/DESm1th/ciftify/archive/v0.1.0.tar.gzTo install with conda, type
conda install -c conda-forge ciftifyAfter completing the previous step, you now need to add ciftify's bin folder to your path. First, locate where the package was installed.
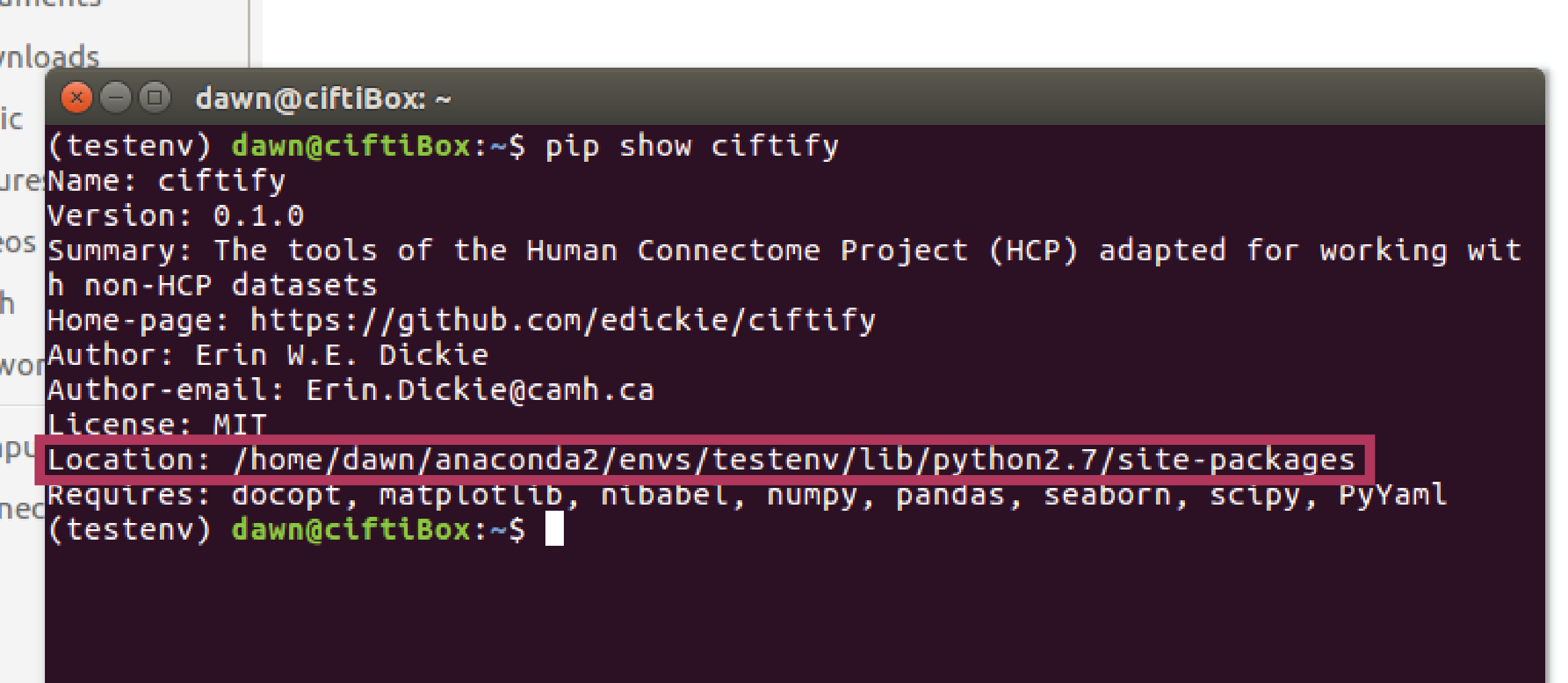
If you installed ciftify with pip, running the command 'pip show ciftify' in the terminal will give you some details about ciftify and the 'location' field will show its install location. The location to add to your path will be this path with 'ciftify/bin' appended to the end. For example, based on the image below ciftify's bin folder would be found at /home/dawn/anaconda2/envs/testenv/lib/python2.7/site-packages/ciftify/bin
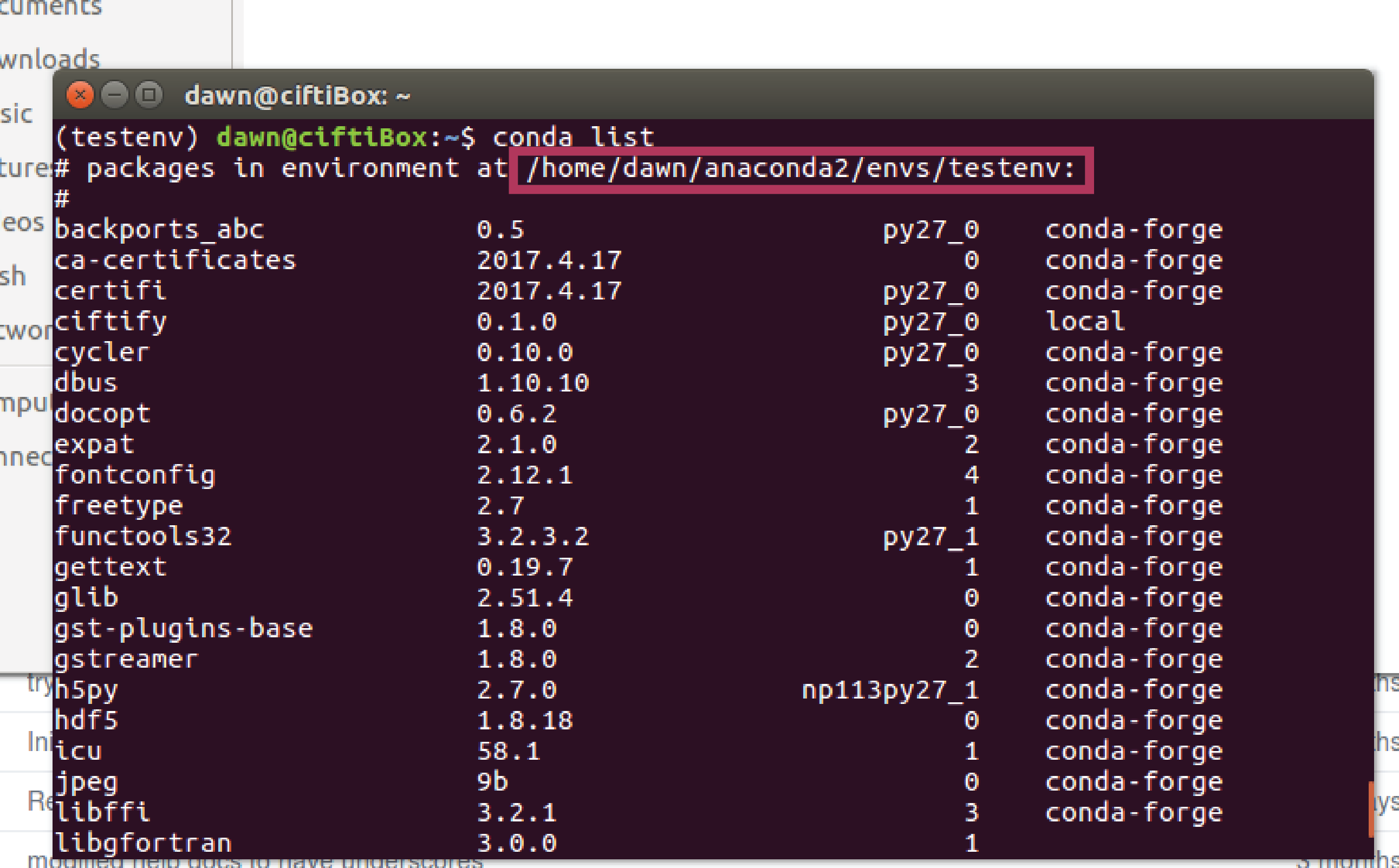
If you installed ciftify with conda, running the command 'conda list' will show you the base path where all packages are installed. To get the exact location of the bin folder from this append 'lib/YOURPYTHONVERSION/site-packages/ciftify/bin'. So, for example, based on the image below and the assumption that python 2.7 is used the bin folder would be found at /home/dawn/anaconda2/envs/testenv/lib/python2.7/site-packages/ciftify/bin
If you don't know your python version for sure you can type python --version in the terminal to check it.
Next, add this location permanently to your PATH by modifying your .bashrc file. The following command will accomplish this. Replace $yourpath with the path pointing to ciftify's bin folder on your computer.
echo 'export PATH="$yourpath:$PATH"' >> ~/.bashrcTo check if ciftify is correctly configured, open a new terminal and type in a ciftify command (like ciftify_a_nifti). If the terminal prints the command's help string you're good to go! Otherwise if the terminal gives you an error like 'command not found', and your spelling of the command was correct, the path you provided has an error in it.
First clone the ciftify repo. Then set some environment variables:
- add the
ciftify/binto yourPATH - add the
ciftifydirectory to yourPYTHONPATH - create a new environment variable (
HCP_SCENE_TEMPLATES) that point to the location of the template scene files - create a new environment variable (
CIFTIFY_TEMPLATES) that points to the location of the data directory. - create an environment variable for the location of your
HCP_DATA
Lastly, install the python package dependencies listed in the 'requirements' section.
MYBASEDIR=${HOME}/code ## change this to the directory you want to clone/download the ciftify code into
cd ${MYBASEDIR}
git clone https://github.com/edickie/ciftify.git
export PATH=$PATH:${MYBASEDIR}/ciftify/bin
export PYTHONPATH=$PYTHONPATH:${MYBASEDIR}/ciftify
export HCP_SCENE_TEMPLATES=${MYBASEDIR}/ciftify/data/scene_templates
export CIFTIFY_TEMPLATES=${MYBASEDIR}/ciftify/data
### optional: you can also set an environment variable to the location of your data
export HCP_DATA=/path/to/hcp/subjects/data/ciftify draws upon the tools and templates of the HCP minimally processed pipelines and therefore is dependent on them and their prereqs:
- HCP Minimal Processing Pipeline (any release) [https://github.com/Washington-University/Pipelines/releases]
- connectome-workbench (tested with version 1.1.1) [http://www.humanconnectome.org/software/get-connectome-workbench]
- FSL [http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/]
- FreeSurfer [https://surfer.nmr.mgh.harvard.edu/fswiki]
- ImageMagick (for cifti-vis image manipultion)
ciftify is mostly written in python 2 (although we believe we are now python 3 compatible!) with the following package dependencies:
- docopt
- matplotlib
- nibabel
- numpy
- pandas
- pyyaml
- seaborn (only for PINT vis)
- scipy
Scripts adapted from HCP Minimal processing pipeline to put preprocessed T1 and fMRI data into an HCP like folder structure
- fs2hcp
- Will convert any freeserfer output directory into an HCP (cifti space) output directory
- func2hcp
- Will project a nifti functional scan to a cifti .dtseries.nii in that subjects hcp analysis directory
- The subject's hcp analysis directory is created by runnning fs2hcp on that participants freesurfer output
- will do fancy outlier removal to optimize the mapping in the process and then smooth the data in cifti space
- cifity_a_nifti
- Will project a nifti scan to cifti space (4D nifti -> .dtseries.nii or 3D nifti -> .dsclar.nii) with no fancy steps or smoothing
- intended for conversion of 3D statistical maps (or 3D regions of interest) for visualization with wb_view
- ciftify_meants:
- extracts mean timeseries(es) (similar to FSL' fslmeants) that can take nifti, cifti or gifti inputs
- ciftify_seed_corr:
- builds seed-based correlation maps using cifti, gifti or nifti inputs
- ciftify_peaktable:
- similar to FSL's clusterize, outputs a csv table of peak locations from a cifti statisical map
- ciftify_surface_rois:
- a tool for building circular rois on the cortical surface. Multiple roi locations can be read at once from a csv table.
- ciftify_groupmask:
- a tools for building a group mask for statiscal analyses using multiple .dtseries.nii files as the input
- citfi_vis_qc:
- builds visual qc pages for verification of fs2hcp and func2hcp conversion
- Note: these pages can also be used for qc of freesurfer's recon-all pipeline
- (they easier to generate (i.e. no display needed) than freesurfer QAtools, and a little prettier too)
- cifti_vis_map:
- generates picture of standard views from any cifti map (combined into on .html page)
- One can loop over multiple files (i.e. maps from multiple subjects) and combine all outputs so that all subjects can viewed together in one index page.
- can also take a nifti input which is internally converted to cifti using ciftify_a_nifti
- cifti_vis_RSN:
- From a functional file input, Will run seed-based correlations from 4 ROIS of interest then generate pics of standard views
- One can loop over multiple files (i.e. maps from multiple subjects) and combine all outputs so that all subjects can viewed together in one index page.
- can also take a nifti input which is internally converted to cifti using ciftify_a_nifti
These two are part of a work in progress (something I need to validate first) ciftify_PINT_vertices cifti_vis_PINT epi_hcpexport