Kian Kelly
library(GGally)
library(tidyverse)
library(scales)
library(network)
library(vroom)
library(phyloseq)
library(vegan)
library(patchwork)
library(ggnewscale)
library(ggforce)
library(geosphere)
library(rnaturalearth)
library(sf)
library(rnaturalearthdata)
library(dplyr)
library(raster)
library(purrr)
library(USAboundaries)
library(USAboundariesData)
library(ggspatial)The TerraClimate project is basically a dataset containing climate station data which was interpolated on a global scale. It was the highest resolution dataset of it’s kind.
#--- Download the files from the TerraClimate website ---#
# Precipitation
if (!file.exists("./ppt.nc")) {
download.file(url = "http://thredds.northwestknowledge.net:8080/thredds/fileServer/TERRACLIMATE_ALL/data/TerraClimate_ppt_2023.nc",
destfile = "ppt.nc")
}
# Evapotranspiration
if (!file.exists("./pet.nc")) {
download.file(url = "http://thredds.northwestknowledge.net:8080/thredds/fileServer/TERRACLIMATE_ALL/data/TerraClimate_pet_2019.nc",
destfile = "pet.nc")
}These are netCDF files which are basically like a CSV but with multiple dimensions (ie climate data for multiple months). Here you can see the raster data for each month of the year. Rasters are basically objects containing values associated with coordinates like x and y or lat and long
#--- Import the downloaded files ---#
# Precipitation
ppt <- stack(x = "ppt.nc")## Loading required namespace: ncdf4
## Warning in .getCRSfromGridMap4(atts): cannot process these parts of the crs:
## long_name=crs
# Evapotranspiration
pet <- stack(x = "pet.nc")## Warning in .getCRSfromGridMap4(atts): cannot process these parts of the crs:
## long_name=crs
#--- Inspect ---#
# Precipitation
plot(ppt)# Evapotranspiration
plot(pet)#--- Raster maths ---#
# Precipitation
ppt_mean <- calc(ppt, # RasterStack object
fun = mean, # Function to apply across the layers
na.rm = TRUE)
# Evapotranspiration
pet_mean <- calc(pet,
fun = mean,
na.rm = TRUE)#--- Set the extent ---#
# Cut off all values outside California
ext <- extent(c(xmin = -124, xmax = -114, ymin = 32, ymax = 42))
#--- Crop ---#
# Precipitation
ppt_mean_cal <- crop(x = ppt_mean, y = ext)
# Evapotranspiration
pet_mean_cal <- crop(x = pet_mean, y = ext)
#--- Inspect ---#
# Precipitation
plot(main = "Precipitation", ppt_mean_cal)# Evapotranspiration
plot(main = "Evapotranspiration", pet_mean_cal)TerraClimate does not automatically calculate aridity. Aridity is the ratio of Precipitation to evapotransperation
#--- Calculate aridity index ---#
# Precipitation (ppt) / Evapotranspiration (pet)
aridity_index <- overlay(x = ppt_mean_cal, # Raster object 1
y = pet_mean_cal, # Raster object 2
fun = function(x, y){return(x / y)}) # Function to apply
plot(main = 'Aridity index',
aridity_index)Here we convert the raster to a dataframe and log normalize for later plotting steps. I am using relative aridity here since this is better for high resolution visualization.
#--- Convert raster to a matrix ---#
aridity_index_matrix <- rasterToPoints(aridity_index)
#--- Convert to the matrix to a dataframe ---#
aridity_index_df <- as.data.frame(aridity_index_matrix)
aridity_index_df$layer = log10(aridity_index_df$layer + 1)You can use USAboundaries package to obtain the California geometry as I did here. If you are interested in another state you can modify this. the geometry is loaded in st format which is not compatible with ggplot so you can use st as sf to convert it.
# Get the geometry of California
ca <- us_states(states = "CA", resolution = "high")
ca_geom <- st_as_sf(ca)You can obtain these shapefiles for different climates using www.sciencebase.gov if you are intrested in adding park borders, etc.
# Path to your shapefile source:
# https://www.sciencebase.gov/catalog/item/5835e1cae4b0d9329c801b7b
shapefile_path_moja <- "./ca_sections.shp"
# Read the shapefile
shapefile_mojave <- st_read(shapefile_path_moja)## Reading layer `ca_sections' from data source
## `/rhome/kkell060/California_Desert_Aridity_Map/ca_sections.shp'
## using driver `ESRI Shapefile'
## Simple feature collection with 5 features and 2 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -2049212 ymin: 1242364 xmax: -1646663 ymax: 2156443
## Projected CRS: USA_Contiguous_Albers_Equal_Area_Conic_USGS_version
# Now you can work with the shapefile object For example,
# you can view its attributes, plot it, etc.
head(shapefile_mojave)## Simple feature collection with 5 features and 2 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -2049212 ymin: 1242364 xmax: -1646663 ymax: 2156443
## Projected CRS: USA_Contiguous_Albers_Equal_Area_Conic_USGS_version
## SECTION HECTARES geometry
## 1 Mojave Desert 6683364.7 MULTIPOLYGON (((-1881344 17...
## 2 Sonoran Desert 1287772.2 MULTIPOLYGON (((-1647076 14...
## 3 Mono 798113.1 MULTIPOLYGON (((-2007430 21...
## 4 Colorado Desert 1185214.8 MULTIPOLYGON (((-1706593 12...
## 5 Southeastern Great Basin 1103856.7 MULTIPOLYGON (((-1913060 18...
# Define the number of colors you want in the palette
num_colors <- 10000
# Generate the palette from white to red
palette <- colorRampPalette(c("beige", "forestgreen", "darkgreen"))(num_colors)the tricky part here is loading in the shapefiles. geom_sf lets you do this. geom_raster allows you to plot the aridity data. How this works is each object has associated coordinates which can be overlayed.
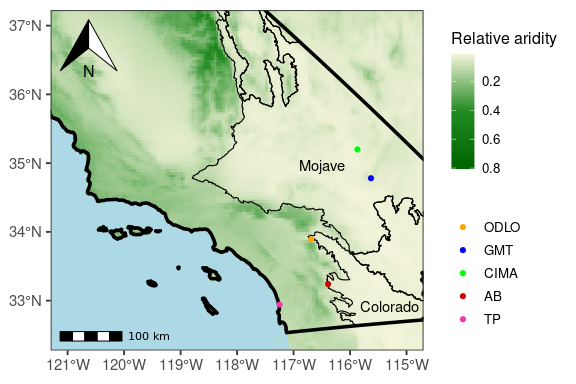
p <- ggplot() +
geom_raster(data = aridity_index_df,
aes(y = y, x = x, fill = layer)) +
scale_fill_gradientn("Relative aridity", colours = palette) +
theme_bw(base_size = 14) +
theme(legend.title = element_blank(),
legend.text = element_text(size = 10),
axis.title = element_blank(),
panel.grid.major = element_line(linetype = 2,
size = 0.5,
colour = 'lightblue'),
panel.grid.minor = element_blank(),
# Set background color to blue
panel.background = element_rect(fill = "lightblue")) +
geom_sf(data = ca_geom, aes(), alpha = 0, colour = "black", linewidth = 1.25) +
geom_sf(data = shapefile_mojave, aes(), alpha = 0, colour = "black", linewidth = .4) +
coord_sf(ylim = c(32.5, 37),
xlim = c(-121, -115)) +
guides(fill = guide_colourbar(reverse = TRUE, title = "Relative aridity",
title.position = "top",
title.theme = element_text(size = 12)),
colour = guide_legend(reverse = TRUE, title = "Relative aridity", title.position = "top")) ## Warning: The `size` argument of `element_line()` is deprecated as of ggplot2 3.4.0.
## ℹ Please use the `linewidth` argument instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
pHere I just made a simple excel sheet with X and Y coordinates for Lat and Long for each of my sites to plot coordinates. I also use ggspatial to add a compas and scalebar.
meta <- read_tsv("./Site_metadata.tsv")## Rows: 5 Columns: 4
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## chr (2): Site, Climate
## dbl (2): Longitude, Lattitude
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
meta$Site <- factor(meta$Site, levels = c("TP", "AB", "CIMA",
"GMT", "ODLO"))
p + ggspatial::annotation_scale(location = "bl", bar_cols = c("black",
"white"), text_family = "ArcherPro Book") + ggspatial::annotation_north_arrow(location = "tl",
which_north = "true", pad_x = unit(0.1, "in"), pad_y = unit(0.1,
"in"), style = ggspatial::north_arrow_orienteering(fill = c("black",
"white"), line_col = "black", )) + geom_text(x = -116.5,
y = 34.8, aes(label = "Mojave"), vjust = -0.5) + geom_text(x = -115.3,
y = 32.75, aes(label = "Colorado"), vjust = -0.5) + geom_point(data = meta,
aes(x = Lattitude, y = Longitude, color = Site)) + scale_color_manual(name = "Geographic Area",
values = c("#F535AA", "red3", "green", "blue", "orange",
"pink"))ggsave(filename = "./art_map.png", plot = last_plot(), device = "png",
width = 10, height = 6, dpi = 300)