Build a comprehensive benchmark of popular Brain-Computer Interface (BCI) algorithms applied on an extensive list of freely available EEG datasets.
This is an open science project that may evolve depending on the need of the community.
First and foremost, Welcome! 🎉 Willkommen! 🎊 Bienvenue! 🎈🎈🎈
Thank you for visiting the Mother of all BCI Benchmark repository.
This document is a hub to give you some information about the project. Jump straight to one of the sections below, or just scroll down to find out more.
- What are we doing? (And why?)
- Installation
- Running
- Supported datasets
- Who are we?
- Get in touch
- Documentation
- Architecture and main concepts
- Citing MOABB and related publications
Brain-Computer Interfaces allow to interact with a computer using brain signals. In this project, we focus mostly on electroencephalographic signals (EEG), that is a very active research domain, with worldwide scientific contributions. Still:
- Reproducible Research in BCI has a long way to go.
- While many BCI datasets are made freely available, researchers do not publish code, and reproducing results required to benchmark new algorithms turns out to be trickier than it should be.
- Performances can be significantly impacted by parameters of the preprocessing steps, toolboxes used and implementation “tricks” that are almost never reported in the literature.
As a result, there is no comprehensive benchmark of BCI algorithms, and newcomers are spending a tremendous amount of time browsing literature to find out what algorithm works best and on which dataset.
The Mother of all BCI Benchmarks allows to:
- Build a comprehensive benchmark of popular BCI algorithms applied on an extensive list of freely available EEG datasets.
- The code is available on GitHub, serving as a reference point for the future algorithmic developments.
- Algorithms can be ranked and promoted on a website, providing a clear picture of the different solutions available in the field.
This project will be successful when we read in an abstract “ … the proposed method obtained a score of 89% on the MOABB (Mother of All BCI Benchmarks), outperforming the state of the art by 5% ...”.
To use MOABB, you could simply do:
pip install MOABB
See Troubleshooting section if you have a problem.
You could fork or clone the repository and go to the downloaded directory, then run:
- install
poetry(only once per machine):
curl -sSL https://install.python-poetry.org | python3 -
or checkout installation instruction or use conda forge version - (Optional, skip if not sure) Disable automatic environment creation:
poetry config virtualenvs.create false - install all dependencies in one command (have to be run in the project directory):
poetry install
See contributors' guidelines for detailed explanation.
See pyproject.toml file for full list of dependencies
To ensure it is running correctly, you can also run
python -m unittest moabb.tests
once it is installed.
First, you could take a look at our tutorials that cover the most important concepts and use cases. Also, we have a several examples available.
You might be interested in MOABB documentation
Moabb has a default image to run the benchmark. You have two options to download this image: build from scratch or pull from the docker hub. We recommend pulling from the docker hub.
If this were your first time using docker, you would need to install the docker and login on docker hub. We recommend the official docker documentation for this step, it is essential to follow the instructions.
After installing docker, you can pull the image from the docker hub:
docker pull baristimunha/moabb
# rename the tag to moabb
docker tag baristimunha/moabb moabbIf you want to build the image from scratch, you can use the following command at the root. You may have to login with the API key in the NGC Catalog to run this command.
bash docker/create_docker.shWith the image downloaded or rebuilt from scratch, you will have an image called moabb.
To run the default benchmark, still at the root of the project, and you can use the
following command:
mkdir dataset
mkdir results
mkdir output
bash docker/run_docker.sh PATH_TO_ROOT_FOLDERAn example of the command is:
cd /home/user/project/moabb
mkdir dataset
mkdir results
mkdir output
bash docker/run_docker.sh /home/user/project/moabbNote: It is important to use an absolute path for the root folder to run, but you can
modify the run_docker.sh script to save in another path beyond the root of the project. By
default, the script will save the results in the project's root in the folder results,
the datasets in the folder dataset and the output in the folder output.
Currently pip install moabb fails when pip version < 21, e.g. with 20.0.2 due to an idna
package conflict. Newer pip versions resolve this conflict automatically. To fix this you
can upgrade your pip version using: pip install -U pip before installing moabb.
The list of supported datasets can be found here : https://neurotechx.github.io/moabb/datasets.html
Detailed information regarding datasets (electrodes, trials, sessions) are indicated on the wiki: https://github.com/NeuroTechX/moabb/wiki/Datasets-Support
you can submit a new dataset by mentioning it to this issue. The datasets currently on our radar can be seen here.
The founders of the Mother of all BCI Benchmarks are Alexander Barachant and Vinay Jayaram. This project is under the umbrella of NeuroTechX, the international community for NeuroTech enthusiasts. The project is currently maintained by Sylvain Chevallier.
You! In whatever way you can help.
We need expertise in programming, user experience, software sustainability, documentation and technical writing and project management.
We'd love your feedback along the way.
Our primary goal is to build a comprehensive benchmark of popular BCI algorithms applied on an extensive list of freely available EEG datasets, and we're excited to support the professional development of any and all of our contributors. If you're looking to learn to code, try out working collaboratively, or translate your skills to the digital domain, we're here to help.
If you think you can help in any of the areas listed above (and we bet you can) or in any of the many areas that we haven't yet thought of (and here we're sure you can) then please check out our contributors' guidelines and our roadmap.
Please note that it's very important to us that we maintain a positive and supportive environment for everyone who wants to participate. When you join us we ask that you follow our code of conduct in all interactions both on and offline.
If you want to report a problem or suggest an enhancement, we'd love for you to open an issue at this GitHub repository because then we can get right on it.
For a less formal discussion or exchanging ideas, you can also reach us on the Gitter channel or join our weekly office hours! This an open video meeting happening on a regular basis, please ask the link on the gitter channel. We are also on NeuroTechX Slack #moabb channel.
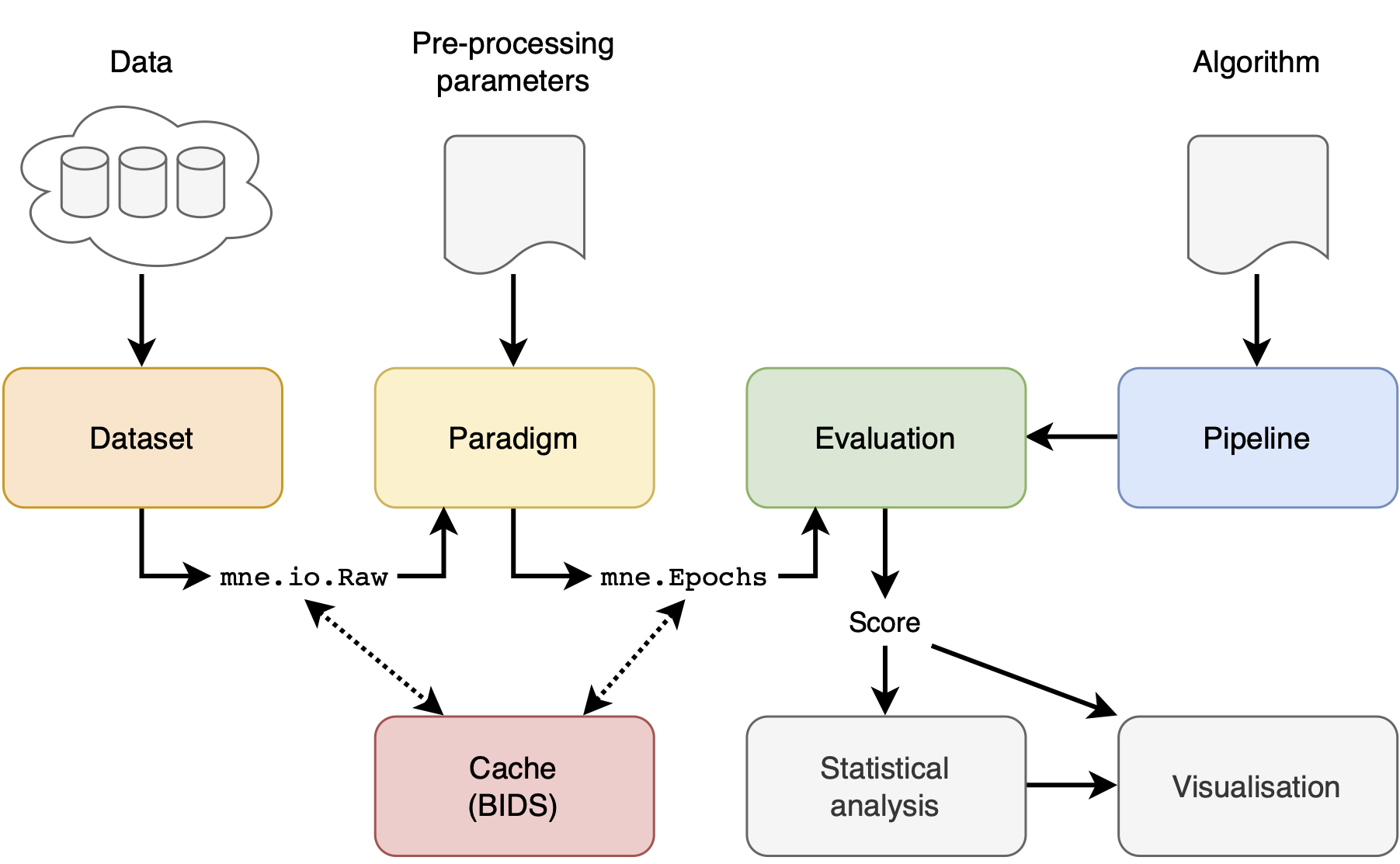
There are 4 main concepts in the MOABB: the datasets, the paradigm, the evaluation, and the pipelines. In addition, we offer statistical and visualization utilities to simplify the workflow.A dataset handles and abstracts low-level access to the data. The dataset will read data stored locally, in the format in which they have been downloaded, and will convert them into a MNE raw object. There are options to pool all the different recording sessions per subject or to evaluate them separately.
A paradigm defines how the raw data will be converted to trials ready to be processed by a decoding algorithm. This is a function of the paradigm used, i.e. in motor imagery one can have two-class, multi-class, or continuous paradigms; similarly, different preprocessing is necessary for ERP vs ERD paradigms.
An evaluation defines how we go from trials per subject and session to a generalization statistic (AUC score, f-score, accuracy, etc) -- it can be either within-recording-session accuracy, across-session within-subject accuracy, across-subject accuracy, or other transfer learning settings.
Pipeline defines all steps required by an algorithm to obtain predictions. Pipelines are typically a chain of sklearn compatible transformers and end with a sklearn compatible estimator. See Pipelines for more info.
Once an evaluation has been run, the raw results are returned as a DataFrame. This can be further processed via the following commands to generate some basic visualization and statistical comparisons:
from moabb.analysis import analyze
results = evaluation.process(pipeline_dict)
analyze(results)
If you use MOABB in your experiments, please cite this library when publishing a paper to increase the visibility of open science initiatives:
Aristimunha, B., Carrara, I., Guetschel, P., Sedlar, S., Rodrigues, P., Sosulski, J., Narayanan, D., Bjareholt, E., Quentin, B., Schirrmeister, R. T.,Kalunga, E., Darmet, L., Gregoire, C., Abdul Hussain, A., Gatti, R., Goncharenko, V., Thielen, J., Moreau, T., Roy, Y., Jayaram, V., Barachant,A., & Chevallier, S. Mother of all BCI Benchmarks (MOABB), 2023. DOI: 10.5281/zenodo.10034223.
and here is the Bibtex version:
@software{Aristimunha_Mother_of_all_2023, author = {Aristimunha, Bruno and Carrara, Igor and Guetschel, Pierre and Sedlar, Sara and Rodrigues, Pedro and Sosulski, Jan and Narayanan, Divyesh and Bjareholt, Erik and Quentin, Barthelemy and Schirrmeister, Robin Tibor and Kalunga, Emmanuel and Darmet, Ludovic and Gregoire, Cattan and Abdul Hussain, Ali and Gatti, Ramiro and Goncharenko, Vladislav and Thielen, Jordy and Moreau, Thomas and Roy, Yannick and Jayaram, Vinay and Barachant, Alexandre and Chevallier, Sylvain}, doi = {10.5281/zenodo.10034223}, title = {{Mother of all BCI Benchmarks}}, url = {https://github.com/NeuroTechX/moabb}, version = {1.0.0}, year = {2023} }
If you want to cite the scientific contributions of MOABB, you could use the following paper:
Vinay Jayaram and Alexandre Barachant. "MOABB: trustworthy algorithm benchmarking for BCIs." Journal of neural engineering 15.6 (2018): 066011. DOI
If you publish a paper using MOABB, please contact us on gitter or open an issue, and we will add your paper to the dedicated wiki page.
Thank you so much (Danke schön! Merci beaucoup!) for visiting the project and we do hope that you'll join us on this amazing journey to build a comprehensive benchmark of popular BCI algorithms applied on an extensive list of freely available EEG datasets.