Component-wise boosting applies the boosting framework to statistical models, e.g., general additive models using component-wise smoothing splines. Boosting these kinds of models maintains interpretability and enables unbiased model selection in high dimensional feature spaces.
The R package compboost is an alternative implementation of
component-wise boosting written in C++ to obtain high runtime
performance and full memory control. The main idea is to provide a
modular class system which can be extended without editing the source
code. Therefore, it is possible to use R functions as well as C++
functions for custom base-learners, losses, logging mechanisms or
stopping criteria.
For an introduction and overview about the functionality visit the project page.
devtools::install_github("schalkdaniel/compboost")This examples are rendered using compboost 0.1.0.
To be as flexible as possible one should use the R6 API do define
base-learner, losses, stopping criteria, or optimizer as desired:
library(compboost)
# Check installed version:
packageVersion("compboost")
## [1] '0.1.0'
# Load data set with binary classification task:
data(PimaIndiansDiabetes, package = "mlbench")
# Create categorical feature:
PimaIndiansDiabetes$pregnant.cat = ifelse(PimaIndiansDiabetes$pregnant == 0, "no", "yes")
# Define Compboost object:
cboost = Compboost$new(data = PimaIndiansDiabetes, target = "diabetes", loss = QuadraticLoss$new())
cboost
## Component-Wise Gradient Boosting
##
## Trained on PimaIndiansDiabetes with target diabetes
## Number of base-learners: 0
## Learning rate: 0.05
## Iterations: 0
## Positive class: neg
##
## QuadraticLoss Loss:
##
## Loss function: L(y,x) = 0.5 * (y - f(x))^2
##
##
# Add p-spline base-learner with default parameter:
cboost$addBaselearner(feature = "pressure", id = "spline", bl.factory = PSplineBlearner)
# Add another p-spline learner with custom parameters:
cboost$addBaselearner(feature = "age", id = "spline", bl.factory = PSplineBlearner, degree = 3,
knots = 10, penalty = 4, differences = 2)
## Warning in .handleRcpp_PSplineBlearner(degree = 3, knots = 10, penalty =
## 4, : Following arguments are ignored by the spline base-learner: knots
# Add categorical feature (as single linear base-learner):
cboost$addBaselearner(feature = "pregnant.cat", id = "category", bl.factory = PolynomialBlearner,
degree = 1, intercept = FALSE)
# Check all registered base-learner:
cboost$getBaselearnerNames()
## [1] "pressure_spline" "age_spline"
## [3] "pregnant.cat_yes_category" "pregnant.cat_no_category"
# Train model:
cboost$train(1000L, trace = FALSE)
cboost
## Component-Wise Gradient Boosting
##
## Trained on PimaIndiansDiabetes with target diabetes
## Number of base-learners: 4
## Learning rate: 0.05
## Iterations: 1000
## Positive class: neg
## Offset: 0.3021
##
## QuadraticLoss Loss:
##
## Loss function: L(y,x) = 0.5 * (y - f(x))^2
##
##
cboost$getBaselearnerNames()
## [1] "pressure_spline" "age_spline"
## [3] "pregnant.cat_yes_category" "pregnant.cat_no_category"
selected.features = cboost$selected()
table(selected.features)
## selected.features
## age_spline pregnant.cat_no_category pressure_spline
## 676 52 272
params = cboost$coef()
str(params)
## List of 4
## $ age_spline : num [1:24, 1] 1.0107 0.5065 0.2452 0.2291 -0.0589 ...
## $ pregnant.cat_no_category: num [1, 1] -0.159
## $ pressure_spline : num [1:24, 1] -0.4856 -0.2185 0.0367 0.2206 0.2616 ...
## $ offset : num 0.302
cboost$train(3000)
##
## You have already trained 1000 iterations.
## Train 2000 additional iterations.
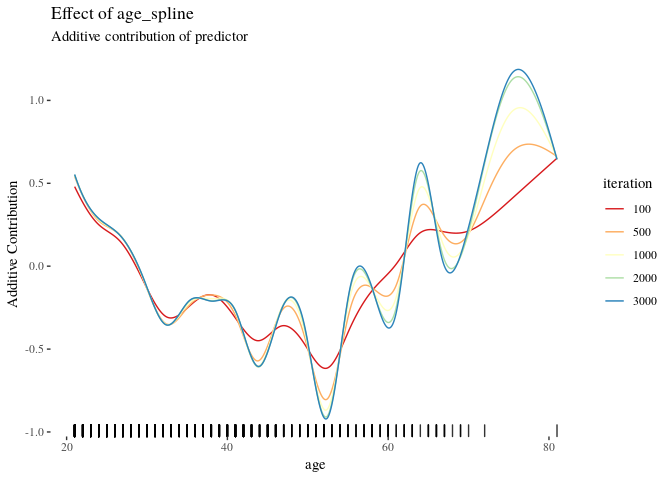
cboost$plot("age_spline", iters = c(100, 500, 1000, 2000, 3000)) +
ggthemes::theme_tufte() +
ggplot2::scale_color_brewer(palette = "Spectral")If you are interested in wrapping each feature, for instance, with a
spline base-learner, then just call the wrapper function
boostSplines():
cboost = boostSplines(data = PimaIndiansDiabetes, target = "diabetes",
loss = BinomialLoss$new(), trace = FALSE)
cboost
## Component-Wise Gradient Boosting
##
## Trained on data with target diabetes
## Number of base-learners: 10
## Learning rate: 0.05
## Iterations: 100
## Positive class: neg
## Offset: 0.3118
##
## BinomialLoss Loss:
##
## Loss function: L(y,x) = log(1 + exp(-2yf(x))
##
##
cboost$getBaselearnerNames()
## [1] "pregnant_spline" "glucose_spline"
## [3] "pressure_spline" "triceps_spline"
## [5] "insulin_spline" "mass_spline"
## [7] "pedigree_spline" "age_spline"
## [9] "pregnant.cat_yes_category" "pregnant.cat_no_category"© 2018 Daniel Schalk
The contents of this repository are distributed under the MIT license. See below for details:
The MIT License (MIT)
Copyright (c) 2018 Daniel Schalk
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.