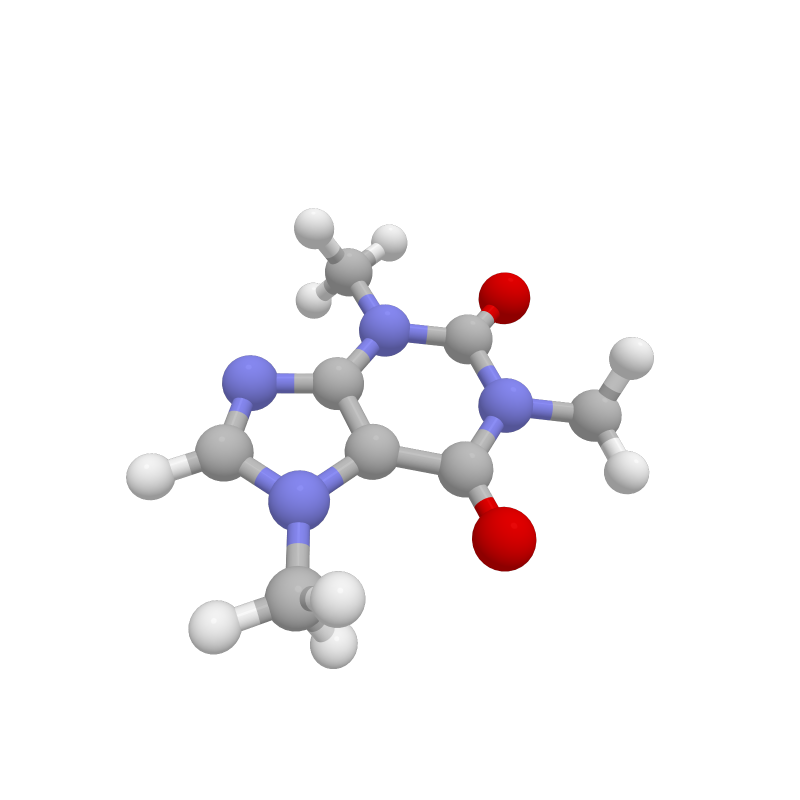
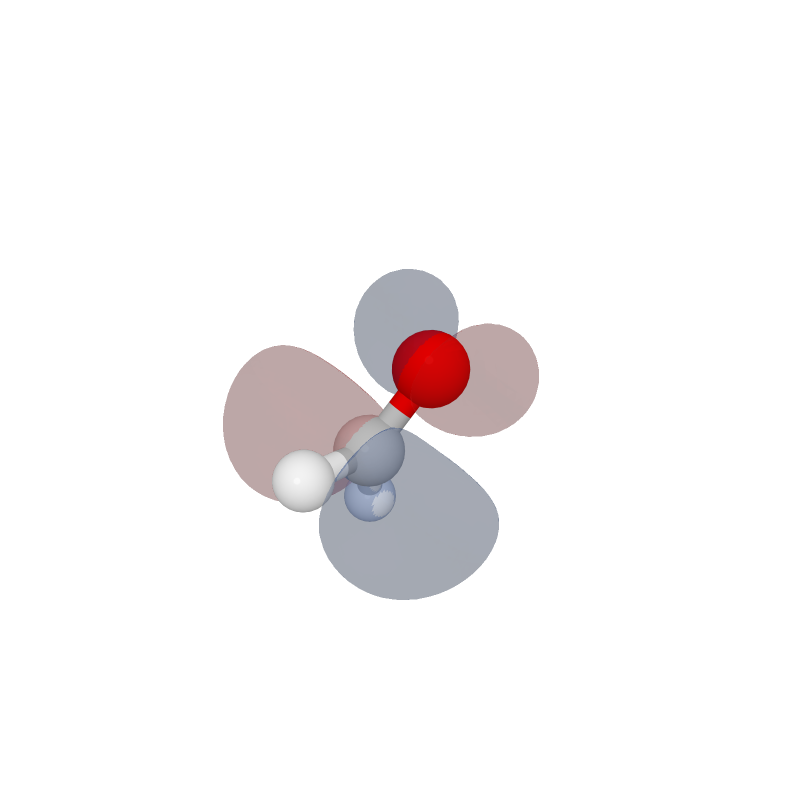
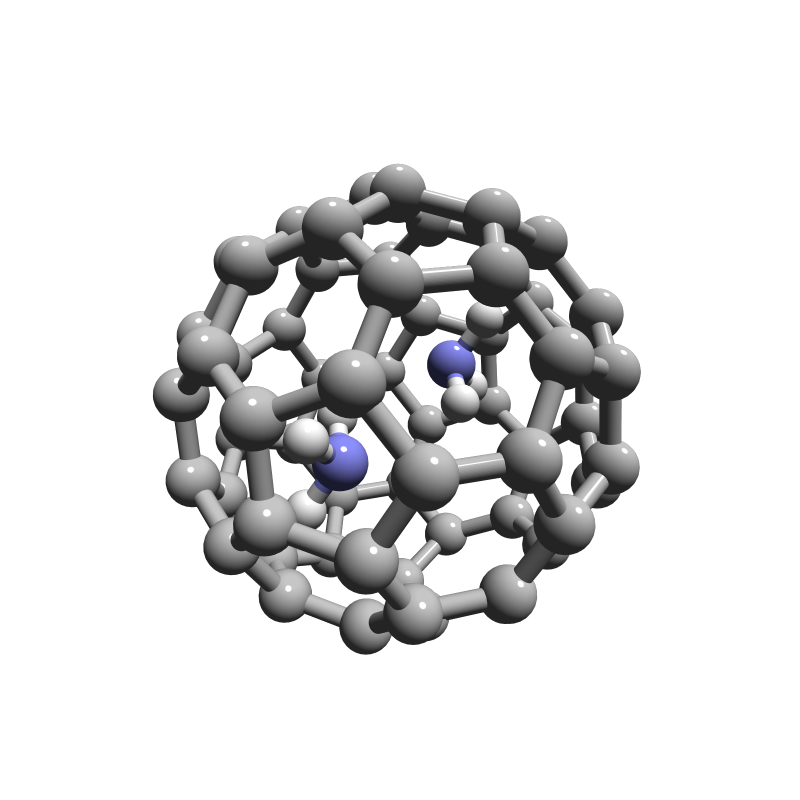
Molecular visualization in Jupyter.

(Package is under development!)
Geometry
Volumes (Density, MOs, ESP)
xyz files
Psi4 geometries
QCElemental molecules
Cube files
- git:
git clone https://github.com/VHchavez/moly.git cd moly pip install . #Be sure to use if release is several commits behind - pip:
pip install moly - conda:
coming soon!
Define a figure and add molecules to it.
import moly
caffeine = moly.molecule_factory("xyz", file='/caffeine.xyz')
fig = moly.Figure()
fig.add_molecule(caffeine)
fig.show()
Geometry and volumentric information can extracted from cube files.
formal = moly.molecule_factory("Cube", file='orbitals.cube')
fig = moly.Figure()
fig.add_molecule(formal)
fig.add_blob(iso=0.05)
fig.show()
Geometries can be brought from different sources and be thrown in the same figure
#Implementation with QCArchive
import qcportal as ptl
client = ptl.FractalClient()
#Get molecule from QCArchive
ds = client.get_collection("ReactionDataset", "S22")
dimers = ds.get_molecules()
ammonia_dimer = dimers.loc['Ammonia Dimer', 'molecule'][0]
#Molecules
ammonia_dimer = moly.molecule_factory("QC", molecule=ammonia_dimer)
bucky = moly.molecule_factory("xyz", file='bucky.xyz')
#Figure, resolution and surface material can be changed
fig = moly.Figure(resolution=(800,800), surface="shiny")
fig.add_molecule(ammonia_dimer)
fig.add_molecule(bucky)
fig.show()
Copyright (c) 2020, VH Chavez
Project based on the Computational Molecular Science Python Cookiecutter version 1.1.