Please see our blog on the IntelAI website
Trains a 2D U-Net on the brain tumor segmentation (BraTS) subset of the Medical Segmentation Decathlon dataset.
Steps:
-
Go to the Medical Segmentation Decathlon website and download the BraTS subset. The dataset has the Creative Commons Attribution-ShareAlike 4.0 International license.
-
Untar the "Task01_BrainTumour.tar" file (e.g.
tar -xvf Task01_BrainTumour.tar) -
We use conda virtual environments to run Python scripts. Once you download and install conda, create a new conda environment with TensorFlow* with Intel® MKL-DNN. Run the command:
conda create -c anaconda -n decathlon pip python=3.7 tensorflow tqdm psutil jupyter matplotlib
This has been tested with TensorFlow 2.2 on Ubuntu 18.04 Linux.
- Enable the new environment. Command:
conda activate decathlon
- Install the package nibabel. Command:
pip install nibabel
- Run the command
bash run_brats_model.sh $DECATHLON_ROOT_DIRECTORY
where $DECATHLON_ROOT_DIRECTORY is the root directory where you un-tarred the Decathlon dataset.
- The bash script should pre-process the Decathlon 3D scan data and store the slices of the 3D Nifti files into separate 2D NumPy files (
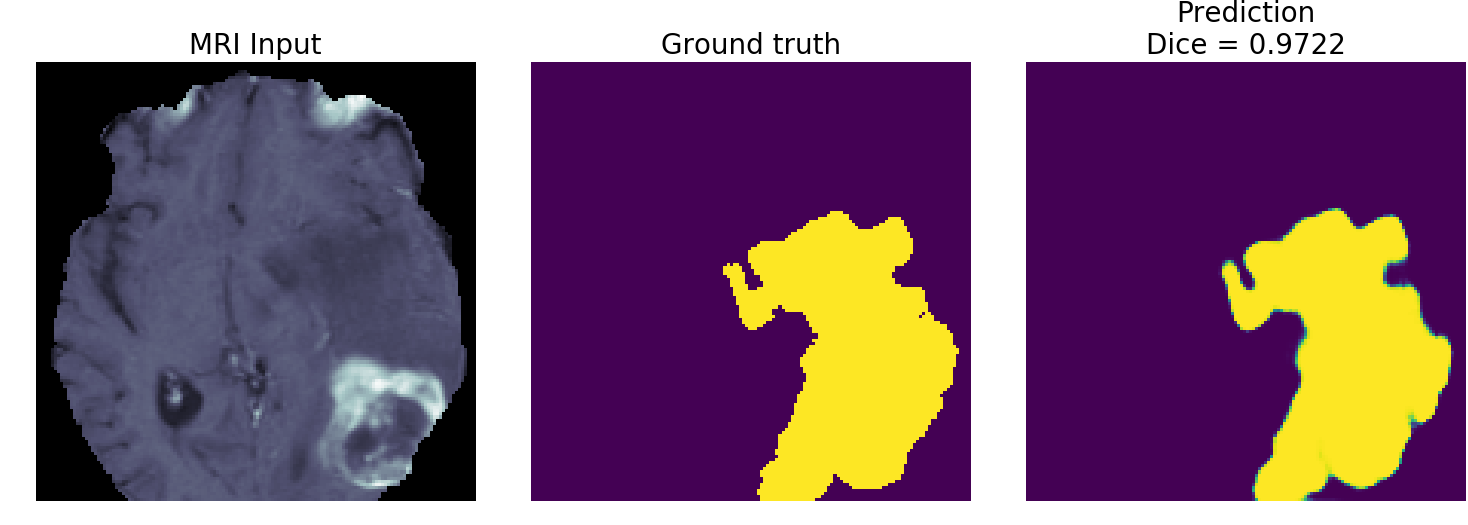
convert_raw_to_npy.py). Then it trains a U-Net model (train.py). Finally, it performs inference on a handful of MRI slices in the validation dataset (plot_tf_inference_examples.py). You should be able to get a model to train to a Dice of over 0.85 on the testing set within 30 epochs.
NOTE: The only reason we convert the 3D files to 2D is to make the data loader easier and faster for the 2D model training. For example, in the 3D model training we simply load the 3D Nifti files directly without this 3D→2D preprocessing step.
- OpenVINO™ - At the end of
train.pyyou should see instructions on how to convert the model for use with the Intel® Distribution of the OpenVINO™ toolkit. Once you have OpenVINO™ installed, you can run a command like the one below to create an OpenVINO™ intermediate representation (IR) of the TensorFlow model. If you are using the Intel® Neural Compute Stick™ (NCS2), simply replace theFP32withFP16in the command below:
source /opt/intel/openvino_2021/bin/setupvars.sh
python $INTEL_OPENVINO_DIR/deployment_tools/model_optimizer/mo_tf.py \
--saved_model_dir ./output/2d_unet_decathlon \
--input_shape [1,128,128,4] \
--model_name 2d_unet_decathlon \
--output_dir ./output/FP32 \
--data_type FP32
This has been tested with the Intel® Distribution of the OpenVINO™ toolkit 2021.2.
- Once you have the OpenVINO™ IR model, you can run the command:
python plot_openvino_inference_examples.py --device CPU
It should give you the same output as the plot_tf_inference_examples.py but execute faster on the same CPU. You can try the options --device GPU or --device=MYRIAD if you have the Intel® integrated GPU or Intel® Neural Compute Stick™ (NCS2) installed on your computer.
For a complete demo showing the Intel® Neural Compute Stick™ (NCS2) try out the Intel® DevCloud for the Edge. You'll be able to try running inference on lots of Intel® hardware using the same OpenVINO™ pipeline.
Tips for improving model:
- The feature maps have been reduced so that the model will train using under 12GB of memory. If you have more memory to use, consider increasing the feature maps using the commandline argument
--featuremaps. The results I plot in the images subfolder are from a model with--featuremaps=32. This will increase the complexity of the model (which will also increase its memory footprint but decrease its execution speed). - If you choose a subset with larger tensors (e.g. liver or lung), it is recommended to add another maxpooling level (and corresponding upsampling) to the U-Net model. This will of course increase the memory requirements and decrease execution speed, but should give better results because it considers an additional recepetive field/spatial size.
- Consider different loss functions. The default loss function here is a weighted sum of
-log(Dice)andbinary_crossentropy. Different loss functions yield different loss curves and may result in better accuracy. However, you may need to adjust thelearning_rateand number of epochs to train as you experiment with different loss functions. The commandline argument--weight_dice_lossdefines the weight to each loss function (loss = weight_dice_loss * -log(dice) + (1-weight_loss_dice)*binary_cross_entropy_loss).
REFERENCES:
-
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Γ, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. "The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)", IEEE Transactions on Medical Imaging 34(10), 1993-2024 (2015) DOI: 10.1109/TMI.2014.2377694
-
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. "Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features", Nature Scientific Data, 4:170117 (2017) DOI: 10.1038/sdata.2017.117
-
Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B, Ronneberger O, Summers RM, Bilic P, Christ PF, Do RKG, Gollub M, Golia-Pernicka J, Heckers SH, Jarnagin WR, McHugo MK, Napel S, Vorontsov E, Maier-Hein L, Cardoso MJ. "A large annotated medical image dataset for the development and evaluation of segmentation algorithms." https://arxiv.org/abs/1902.09063
Please see our optimization notice.