24 June, 2020
- Load datasets
- Preprocessing
- Issue I: Absolute vs relative values
- Issue II: Dates vs smart reference date
- Issue III: Infected vs active cases
- Issue IV: This is the past
#'
#' Load COVID-19 spread: infected, recovered, and fatal cases
#' Source: https://github.com/CSSEGISandData/COVID-19/tree/master/csse_covid_19_data/csse_covid_19_time_series
#'
load_covid_spread <- function() {
require(dplyr)
require(data.table)
require(purrr)
require(tidyr)
load_time_series <- function(.case_type) {
path_pattern <- "https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_%s_global.csv"
fread(sprintf(path_pattern, .case_type)) %>%
rename(country = `Country/Region`) %>%
select(-c(`Province/State`, Lat, Long)) %>%
group_by(country) %>%
summarise_if(is.numeric, sum) %>%
ungroup %>%
gather(key = "date", value = "n", -country) %>%
mutate(date = mdy(date))
}
dt <- load_time_series("confirmed") %>% rename(confirmed_n = n) %>%
inner_join(
load_time_series("recovered") %>% rename(recovered_n = n),
by = c("country", "date")
) %>%
inner_join(
load_time_series("deaths") %>% rename(deaths_n = n),
by = c("country", "date")
)
stopifnot(nrow(dt) > 0)
return(dt)
}
spread_raw <- load_covid_spread()## Loading required package: data.table
##
## Attaching package: 'data.table'
## The following objects are masked from 'package:xts':
##
## first, last
## The following objects are masked from 'package:lubridate':
##
## hour, isoweek, mday, minute, month, quarter, second, wday, week, yday, year
## The following object is masked from 'package:purrr':
##
## transpose
## The following objects are masked from 'package:dplyr':
##
## between, first, last
spread_raw %>% sample_n(10)## # A tibble: 10 x 5
## country date confirmed_n recovered_n deaths_n
## <chr> <date> <int> <int> <int>
## 1 Central African Republic 2020-02-23 0 0 0
## 2 Australia 2020-04-07 5895 1080 45
## 3 Panama 2020-02-23 0 0 0
## 4 Kuwait 2020-02-04 0 0 0
## 5 Bangladesh 2020-02-03 0 0 0
## 6 Cuba 2020-05-13 1810 1326 79
## 7 Costa Rica 2020-03-09 9 0 0
## 8 Israel 2020-01-30 0 0 0
## 9 Greece 2020-02-04 0 0 0
## 10 Taiwan* 2020-06-05 443 429 7
#'
#' Load countries stats
#' Source: https://ods.ai/competitions/sberbank-covid19-forecast
#'
load_countries_stats <- function() {
require(dplyr)
require(magrittr)
dt <- fread("https://raw.githubusercontent.com/codez0mb1e/covid-2019/master/data/countries.csv")
dt %<>%
select(-c(iso_alpha2, iso_numeric, name, official_name))
stopifnot(nrow(dt) > 0)
return(dt)
}
countries_raw <- load_countries_stats()
countries_raw %>% sample_n(10)## iso_alpha3 ccse_name density fertility_rate land_area median_age migrants population
## 1: DOM Dominican Republic 225 2.4 48320 28 -30000 10847910
## 2: BRN Brunei 83 1.8 5270 32 0 437479
## 3: GUY Guyana 4 2.5 196850 27 -6000 786552
## 4: PER Peru 26 2.3 1280000 31 99069 32971854
## 5: CYP Cyprus 131 1.3 9240 37 5000 1207359
## 6: ESP Spain 94 1.3 498800 45 40000 46754778
## 7: GEO Georgia 57 2.1 69490 38 -10000 3989167
## 8: PHL Philippines 368 2.6 298170 26 -67152 109581078
## 9: PAK Pakistan 287 3.6 770880 23 -233379 220892340
## 10: ETH Ethiopia 115 4.3 1000000 19 30000 114963588
## urban_pop_rate world_share
## 1: 0.85 0.0014
## 2: 0.80 0.0001
## 3: 0.27 0.0001
## 4: 0.79 0.0042
## 5: 0.67 0.0002
## 6: 0.80 0.0060
## 7: 0.58 0.0005
## 8: 0.47 0.0141
## 9: 0.35 0.0283
## 10: 0.21 0.0147
stopifnot(
nrow(spread_raw %>% group_by(country, date) %>% filter(n() > 1)) == 0
)
data <- spread_raw %>%
# add country population
inner_join(
countries_raw %>% transmute(ccse_name, country_iso = iso_alpha3, population) %>% filter(!is.na(country_iso)),
by = c("country" = "ccse_name")
) %>%
# calculate active cases
mutate(
active_n = confirmed_n - recovered_n - deaths_n
) %>%
# calculate cases per 1M population
mutate_at(
vars(ends_with("_n")),
list("per_1M" = ~ ./population*1e6)
)
## Calculte number of days since...
get_date_since <- function(dt, .case_type, .n) {
dt %>%
group_by(country) %>%
filter_at(vars(.case_type), ~ . > .n) %>%
summarise(since_date = min(date))
}
data %<>%
inner_join(
data %>% get_date_since("confirmed_n", 0) %>% rename(since_1st_confirmed_date = since_date),
by = "country"
) %>%
inner_join(
data %>% get_date_since("confirmed_n_per_1M", 1) %>% rename(since_1_confirmed_per_1M_date = since_date),
by = "country"
) %>%
inner_join(
data %>% get_date_since("deaths_n_per_1M", .1) %>% rename(since_dot1_deaths_per_1M_date = since_date),
by = "country"
) %>%
mutate_at(
vars(starts_with("since_")),
list("n_days" = ~ difftime(date, ., units = "days") %>% as.numeric)
) %>%
mutate_at(
vars(ends_with("n_days")),
list(~ if_else(. > 0, ., NA_real_))
)## Note: Using an external vector in selections is ambiguous.
## ℹ Use `all_of(.case_type)` instead of `.case_type` to silence this message.
## ℹ See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
## This message is displayed once per session.
## `summarise()` ungrouping output (override with `.groups` argument)
## `summarise()` ungrouping output (override with `.groups` argument)
## `summarise()` ungrouping output (override with `.groups` argument)
ggplot(data %>% filter_countries, aes(x = date)) +
geom_col(aes(y = confirmed_n), alpha = .9) +
scale_x_date(date_labels = "%d %b", date_breaks = "7 days") +
facet_grid(country ~ .) +
labs(x = "", y = "# of cases",
title = "COVID-19 Spread by Country",
subtitle = "Infected cases over time",
caption = lab_caption) +
theme(plot.caption = element_text(size = 8))ggplot(data %>% filter_countries, aes(x = date)) +
geom_col(aes(y = confirmed_n), alpha = .9) +
scale_x_date(date_labels = "%d %b", date_breaks = "7 days") +
scale_y_log10() +
facet_grid(country ~ .) +
labs(x = "", y = "# of cases",
title = "COVID-19 Spread by Country",
subtitle = "Infected cases over time",
caption = lab_caption) +
theme(plot.caption = element_text(size = 8))ggplot(data %>% filter_countries, aes(x = date)) +
geom_col(aes(y = confirmed_n_per_1M), alpha = .9) +
scale_x_date(date_labels = "%d %b", date_breaks = "7 days") +
facet_grid(country ~ .) +
labs(x = "", y = "# of cases per 1M",
subtitle = "Infected cases per 1 million popultation",
title = "COVID-19 Spread (over time)",
caption = lab_caption) +
theme(plot.caption = element_text(size = 8))ggplot(data %>% filter_countries, aes(x = date)) +
geom_col(aes(y = confirmed_n_per_1M), alpha = .9) +
scale_x_date(date_labels = "%d %b", date_breaks = "7 days") +
scale_y_log10() +
facet_grid(country ~ .) +
labs(x = "", y = "# of cases per 1M",
subtitle = "Infected cases per 1 million popultation (over time)",
title = "COVID-19 Spread by Country",
caption = lab_caption) +
theme(plot.caption = element_text(size = 8))ggplot(
data %>% filter_countries %>% filter(!is.na(since_1st_confirmed_date_n_days)),
aes(x = since_1st_confirmed_date_n_days)
) +
geom_col(aes(y = confirmed_n), alpha = .9) +
scale_y_log10() +
facet_grid(country ~ .) +
labs(x = "# of days since 1st infected case", y = "# of cases",
subtitle = "Infected cases since 1st infected case",
title = "COVID-19 Spread by Country",
caption = lab_caption) +
theme(plot.caption = element_text(size = 8))ggplot(
data %>% filter_countries %>% filter(!is.na(since_1_confirmed_per_1M_date_n_days)),
aes(x = since_1_confirmed_per_1M_date_n_days)
) +
geom_col(aes(y = confirmed_n_per_1M), alpha = .9) +
scale_y_log10() +
xlim(c(0, 100)) +
facet_grid(country ~ .) +
labs(x = "# of days since 1 infected cases per 1M", y = "# of cases per 1M",
title = "COVID-19 Spread by Country",
subtitle = "Since 1 infected cases per 1 million popultation",
caption = lab_caption) +
theme(plot.caption = element_text(size = 8))plot_data <- data %>%
filter_countries %>%
filter(!is.na(since_1_confirmed_per_1M_date_n_days)) %>%
mutate(
confirmed_n_per_1M = confirmed_n_per_1M,
recovered_n_per_1M = -recovered_n_per_1M,
deaths_n_per_1M = -deaths_n_per_1M
) %>%
select(
country, since_1_confirmed_per_1M_date_n_days, ends_with("_n_per_1M")
) %>%
gather(
key = "case_state", value = "n_per_1M", -c(country, since_1_confirmed_per_1M_date_n_days, active_n_per_1M)
)
ggplot(plot_data, aes(x = since_1_confirmed_per_1M_date_n_days)) +
geom_col(aes(y = n_per_1M, fill = case_state), alpha = .9) +
geom_line(aes(y = active_n_per_1M), color = "#0080FF", size = .25) +
scale_fill_manual(element_blank(),
labels = c("confirmed_n_per_1M" = "Infected cases", "recovered_n_per_1M" = "Recovered cases", "deaths_n_per_1M" = "Fatal cases"),
values = c("confirmed_n_per_1M" = "grey", "recovered_n_per_1M" = "gold", "deaths_n_per_1M" = "black")) +
xlim(c(0, 100)) +
facet_grid(country ~ ., scales = "free") +
labs(x = "# of days since 1 infected cases per 1M", y = "# of cases per 1M",
title = "COVID-19 Spread by Countries",
subtitle = "Active cases trend since 1 infected cases per 1 million popultation. \nBlue line - infected cases minus recovered and fatal.\nNegative values indicate recovered and fatal cases.",
caption = lab_caption) +
theme(
legend.position = "top",
plot.caption = element_text(size = 8)
)## Warning: Removed 309 rows containing missing values (position_stack).
## Warning: Removed 12 rows containing missing values (geom_col).
## Warning: Removed 24 row(s) containing missing values (geom_path).
forecast_cases <- function(.country, .after_date, .forecast_horizont, .fun, ...) {
dt <- data %>%
# filter rows and cols
filter(
country == .country &
date < .after_date
) %>%
# convert to time-series
arrange(date) %>%
select(active_n_per_1M)
dt %>%
ts(frequency = 7) %>%
# ARIMA model
.fun(...) %>%
# forecast
forecast(h = .forecast_horizont)
}
forecast_horizont <- 7
after_date <- max(data$date) + days()
countries_list <- c("Belgium", "France", "Italy", "Netherlands", "Norway", "Portugal", "Spain", "Switzerland", "US", "Russia", "China", "Korea, South")
pred <- countries_list %>%
map_dfr(
function(.x) {
m <- forecast_cases(.x, after_date, forecast_horizont, auto.arima)
n_days_max <- max(data[data$country == .x, ]$since_1_confirmed_per_1M_date_n_days, na.rm = T)
tibble(
country = rep(.x, forecast_horizont),
since_1_confirmed_per_1M_date_n_days = seq(n_days_max + 1, n_days_max + forecast_horizont, by = 1),
pred = m$mean %>% as.numeric %>% round %>% as.integer,
data_type = "Forecast"
)
}
)
plot_data <- data %>%
filter(country %in% countries_list) %>%
transmute(
country, active_n_per_1M, since_1_confirmed_per_1M_date_n_days,
data_type = "Historical data"
) %>%
bind_rows(
pred %>% rename(active_n_per_1M = pred)
) %>%
mutate(
double_every_10d = (1 + 1/10)^since_1_confirmed_per_1M_date_n_days, # double every 2 weeks
double_every_7d = (1 + 1/7)^since_1_confirmed_per_1M_date_n_days, # double every week
double_every_3d = (1 + 1/3)^since_1_confirmed_per_1M_date_n_days, # double every 3 days
double_every_2d = (1 + 1/2)^since_1_confirmed_per_1M_date_n_days # double every 2 days
)
active_n_per_1M_last <- plot_data %>%
group_by(country) %>%
arrange(desc(since_1_confirmed_per_1M_date_n_days)) %>%
filter(row_number() == 1) %>%
ungroup
plot_data %<>%
left_join(
active_n_per_1M_last %>% transmute(country, active_n_per_1M_last = active_n_per_1M, since_1_confirmed_per_1M_date_n_days),
by = c("country", "since_1_confirmed_per_1M_date_n_days")
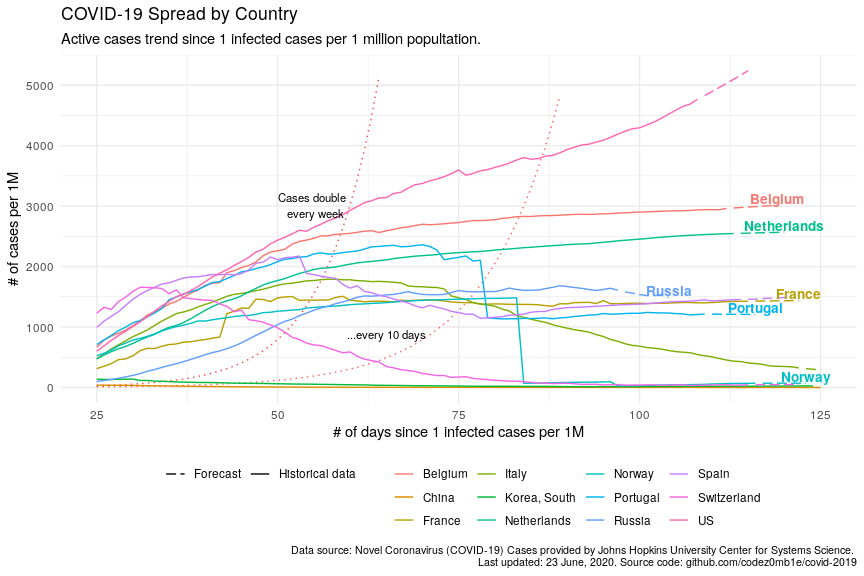
)ggplot(plot_data, aes(x = since_1_confirmed_per_1M_date_n_days)) +
geom_line(aes(y = double_every_10d), linetype = "dotted", color = "red", alpha = .65) +
geom_line(aes(y = double_every_7d), linetype = "dotted", color = "red", alpha = .75) +
geom_line(aes(y = active_n_per_1M, color = country, linetype = data_type), show.legend = T) +
geom_text(aes(y = active_n_per_1M_last + 20, label = country, color = country),
hjust = 0.5, vjust = 0, check_overlap = T, show.legend = F, fontface = "bold", size = 3.6) +
annotate(geom = "text", label = "Cases double \n every week", x = 55, y = 2800, vjust = 0, size = 3) +
annotate(geom = "text", label = "...every 10 days", x = 65, y = 800, vjust = 0, size = 3) +
scale_linetype_manual(values = c("longdash", "solid")) +
xlim(c(25, 125)) +
ylim(c(0, max(plot_data$active_n_per_1M))) +
labs(x = "# of days since 1 infected cases per 1M", y = "# of cases per 1M",
title = "COVID-19 Spread by Country",
subtitle = "Active cases trend since 1 infected cases per 1 million popultation.",
caption = lab_caption) +
theme(
legend.position = "bottom",
legend.title = element_blank(),
plot.caption = element_text(size = 8)
)Take Care and Stay Healthy!