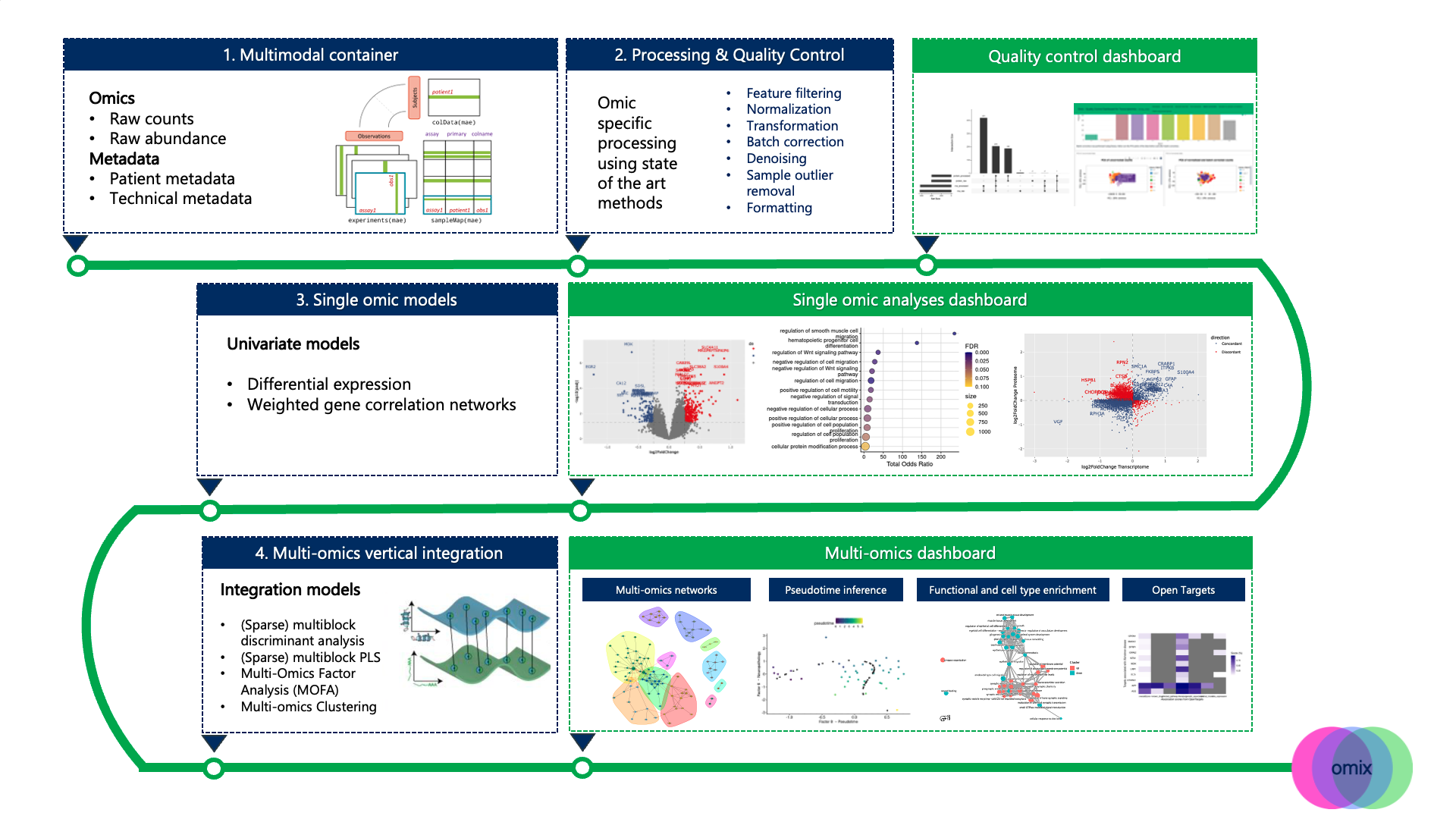
The Omix pipeline provides a generalisable framework that can effectively process, visualise and analyse patient-specific multi-omic data in an end-to-end manner. Omix is built on four consecutive blocks, summarised in the figure below.
This bioinformatics pipeline aims to address important gaps related to the integration of multi-omics data, namely by enabling the reproducibility of integrative analyses and providing grounds for more systematic benchmarking of analyses. Pipeline outputs are standardized and include publication-quality plots, tables, and interactive dashboards.
You can install the development version of Omix from GitHub with:
# install.packages("devtools")
devtools::install_github("eleonore-schneeg/Omix")- Multi-omics data container
- The Omix multimodal container harmonises data management of multiple omics datasets. It enables the storage of raw and processed omics data slots, along with patient metadata, technical metadata, analysis parameters and outputs. The object structure relies on the MultiAssayExperiment library
- Data processing & Quality Control
- Each omics layer is processed separately according to best practices. Given the wide range of processing functionalities, users decide which parameters and steps of the modular sequence are performed, which involves all or a combination of the folllowing steps:
- Feature filtering
- Normalisation/ transformation
- Batch correction & denoising
- Sample outlier removal
- Formatting
- Single platform models
- Omix provides a suite of analysis options including differential analysis (DE), a standard method to identify genes that are differentially expressed between certain disease states; Weighted Gene correlation Network (WGCNA), to identify modules of genes that associate to certain disease covariates; sparse Partial Least Square (sPLS) to define a sparse set of omics features, or molecular signature, that explains the response variable.
- Vertical integration for joint analysis
- Omix provides a series of state-of-the-art integration models to perform patient-specific multi-omic integration, including:
- MOFA
- Sparse Multi-Block PLS (sMB-PLS)
- DIABLO
- A range of multi-omics clustering models implemented in MOVICS
- Downstream analyses
- Multi-omics integration is followed by a series of downstream analyses, including:
- Multi-omics networks with iGraph
- Community detection with the Louvain or Leiden clustering algorithms
- Pseudotime inference with Slingshot
- Functional enrichment with EnrichR
- Cell-type enrichment with EWCE
- Target validation based on the OpenTargets database
- Publication quality plots and analysis reports
Omix implements these modular steps and displays results in an interactive results dashboard.
The package functions are designed to interface neatly with NextFlow for scalable and containerized pipelines deployed locally, on high-performance computing clusters, or in the cloud. See https://nf-co.re/omix for the accompanying NextFlow pipeline.