Baker Moran
This is a package to allow AB testers to create a standard report for test results. Can handle conversion and continuous data. Can handle multiple variants.
Conversion data uses the beta distribution as the conjugate prior, and continuous data uses the log-normal.
raw_data- a pandas dataframe with (at least) two columns, one for the bucket and one for the response variablemetric- column name inraw_datafor the response variablebucket_col_name- column name inraw_datafor the bucket (defaults tobucket)control_bucket_name- value inbucket_col_namefor the control bucket (defaults tooff)samples- number of samples to run the monte carlo simulation, must be 1,000 and 50,000 (defaults to 10,000)prior_func- the type of distribution to use for the prior. options include:beta- use for conversion rates. bounded on the interval [0,1]log-normal- use for continuous, greater than zero response variables (0, +inf) (ex: premium, minutes spent on scene, etc.)normal- use for continuous response variables that on the interval (-inf, +inf)poisson- use for discrete, greater than zero response variables (ex: arrivals per day, events per account, etc.)
prior_info- the prior belief to about the response varable. options include:uninformed- no prior belief about the response, all outcomes are equaly likely. No input required to useinformed- uses and empirically informed prior (informed by the control data), and weakens this prior byprior_scale_factorspecified- allows for a user to input prior parameters
prior_parameters-prior_infomust be set tospecified. This must be a dictionary with the following key value pairs:prior_func=beta- keys arealphaandbetaprior_func=log-normal- keys aremeanandvarprior_func=normal- keys aremeanandvarprior_func=poisson- keys arealphaandbetaORmeanandvar
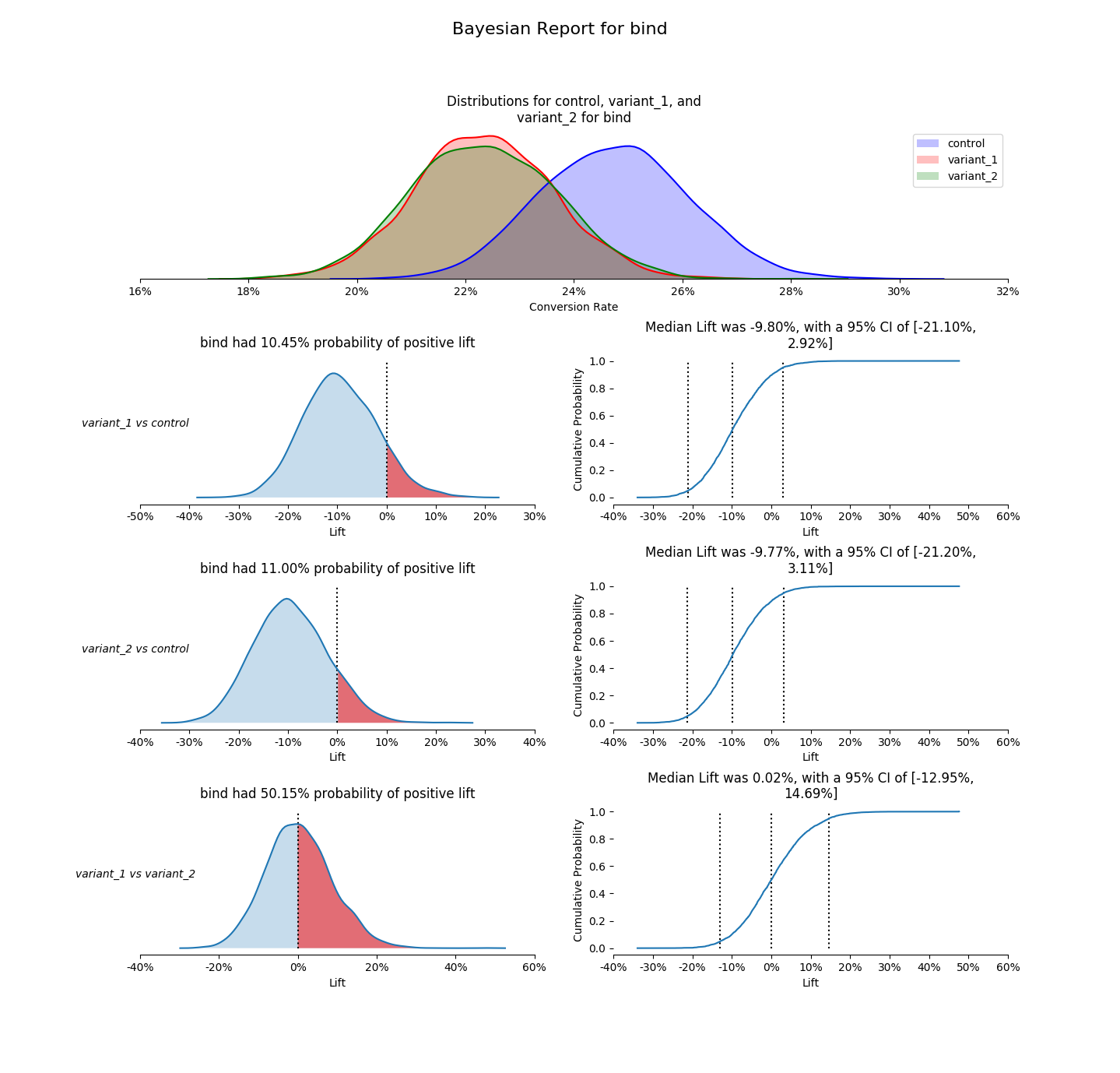
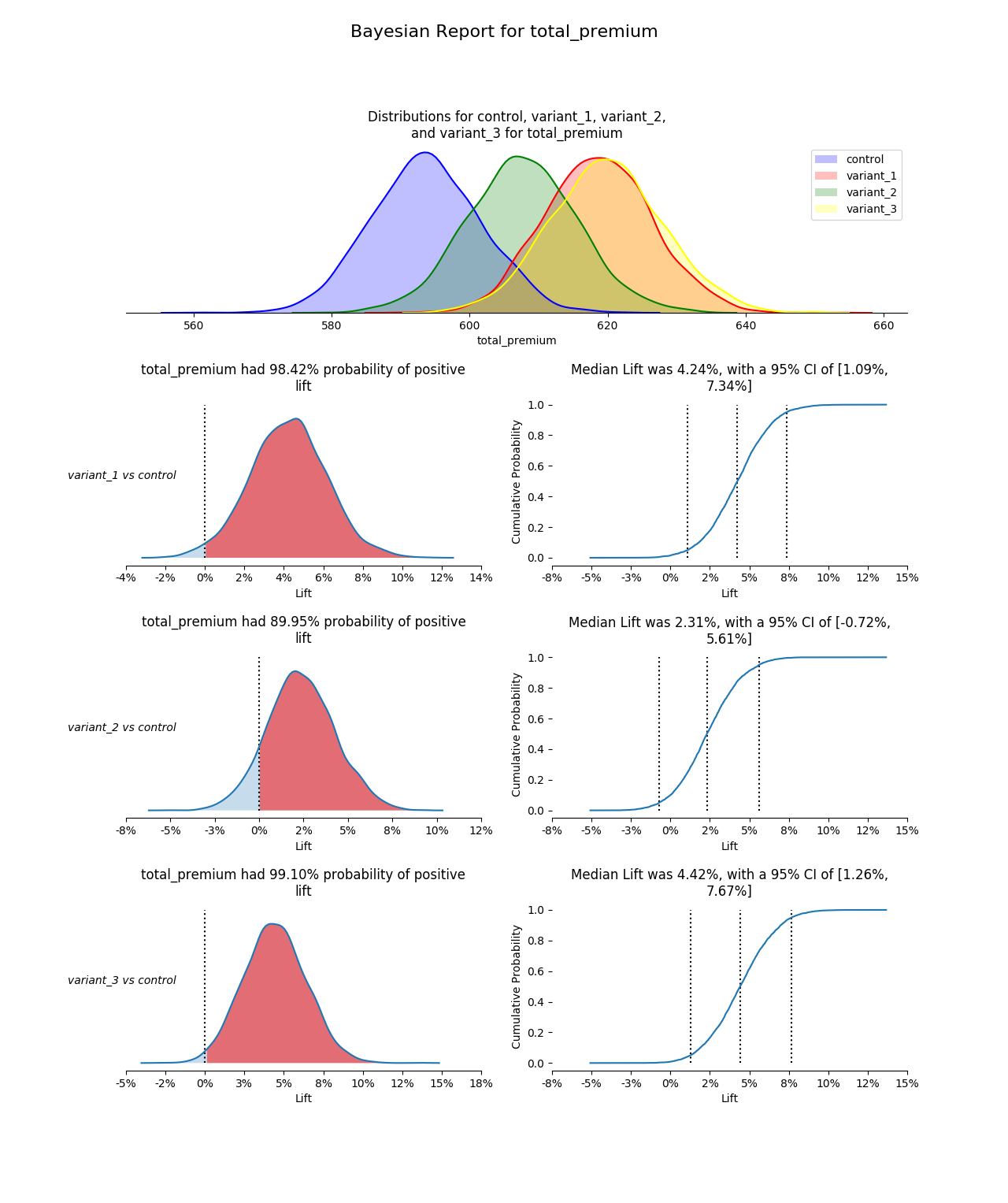
confidence_level- value for the confidence interval on the CDF chart (defaults to0.05)compare_variants- boolean for comparing the variants to each other. Control to each variant is always done (unless there are too many variants to plot). If there are few enough variants, the comparisons for variants will be plotted. (defaults toFalse)debug- boolean to print out extra output for debugging purposes (defaults toFalse)prior_scale_factor- factor to scale an empirically informed prior by (defaults to4)
fit()- requires - class was instantiated with valid inputs
- modifies -
ecdf,control_sample,variant_sample,lift - effects - creates and runs the monte carlo simulation, sets member variables to reflect model outputs
plot(lift_plot_flag=True)lift_plot_flag- boolean for plotting lift PDF and CDF (defaults toTrue)- requires -
fit()has been run - modifies - none
- effects - creates a 3 chart report of the AB test. (must show with
matplotlib.pylot.show())
plot_posteriors(variants=[]):- requires - variants is a list of variant names in
bucket_col_name. One variant may not be repeated multiple times. - modifies - none
- effects - creates a posterior plot of these variants (must show with
matplotlib.pylot.show())
- requires - variants is a list of variant names in
plot_positive_lift(variant_one, variant_two)- requires - variant_one and variant_two are variant names in
bucket_col_name. One variant may not be repeated multiple times. - modifies - none
- effects - creates a positive lift plot between these variants (must show with
matplotlib.pylot.show())
- requires - variant_one and variant_two are variant names in
plot_ecdf(variant_one, variant_two)- requires - variant_one and variant_two are variant names in
bucket_col_name. One variant may not be repeated multiple times. - modifies - none
- effects - creates an empirical cumulative distribution plot of these variants lift (must show with
matplotlib.pylot.show())
- requires - variant_one and variant_two are variant names in
- all variables in
Instantiation variables ecdf- a 3 level dicitonary, where key1 is a the variant name of the numerator in the lift calculation, and key2 is the variant name of the denominator in the lift calculation. The third keys arexandy, and the value is a list containing the x and y coordinates for the empirical CDF (only meaningful afterfit()is run)lift- a 2 level dicitonary, where key1 is a the variant name of the numerator in the lift calculation, and key2 is the variant name of the denominator in the lift calculation. The value is a list containing the lift of the variant over the control for each sampled value (only meaningful afterfit()is run)posteriorsis a dictionary, where the key is a variant name, and the value is aposterior_distributionobject.
First install package. Navigate to the directory in terminal and pip install it.
cd BayesABTest
pip3 install -e .Second, import in your analysis file.
import pandas as pd
from BayesABTest import ab_test_model
# read in data
df = pd.read_csv('some_file.csv')
# initialize class with the data and some optional variables
first_test = ab_test_model(df, metric='bind', prior_info='informed', prior_func='beta', debug=True)
# run public methods
first_test.fit()
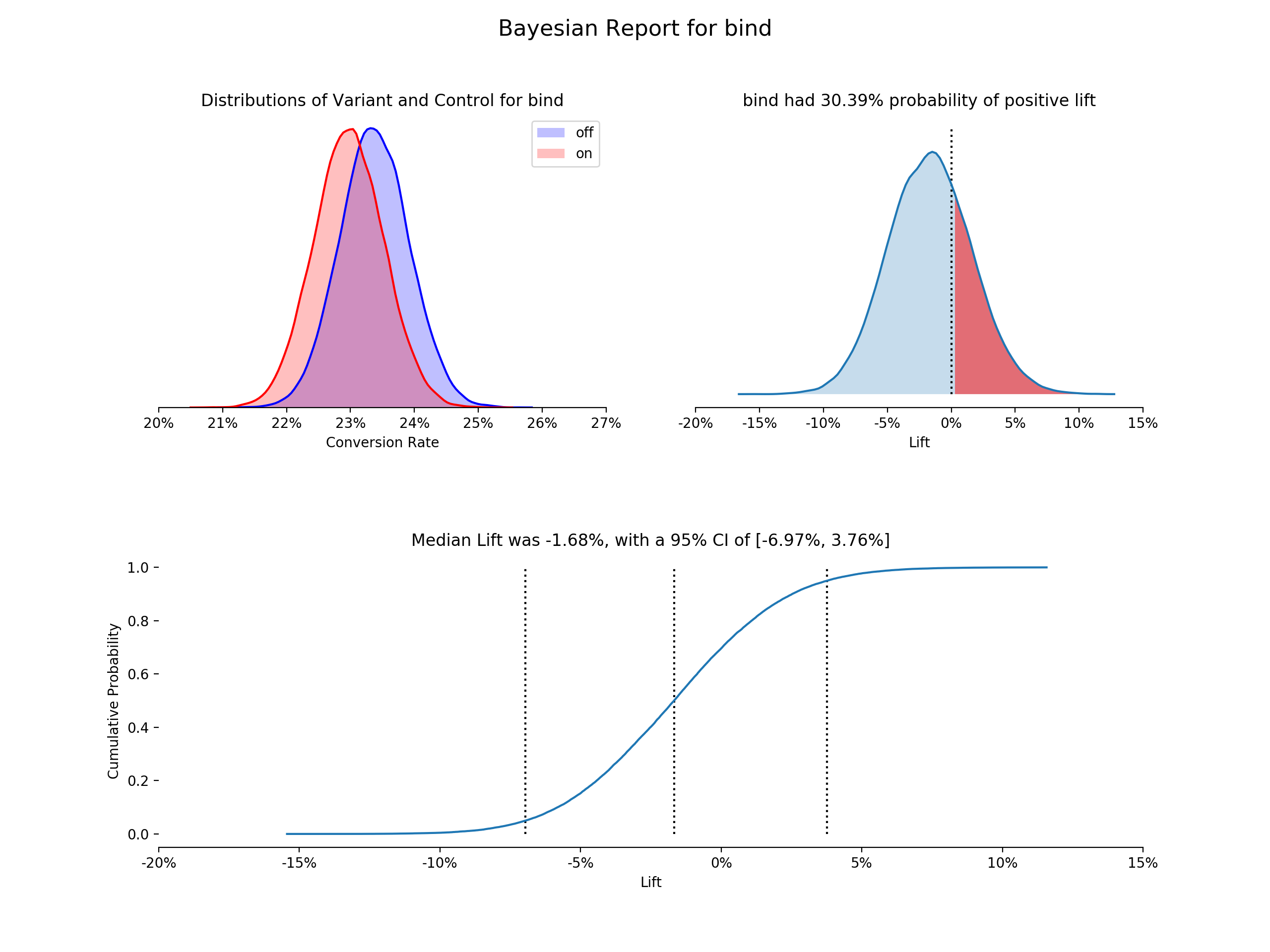
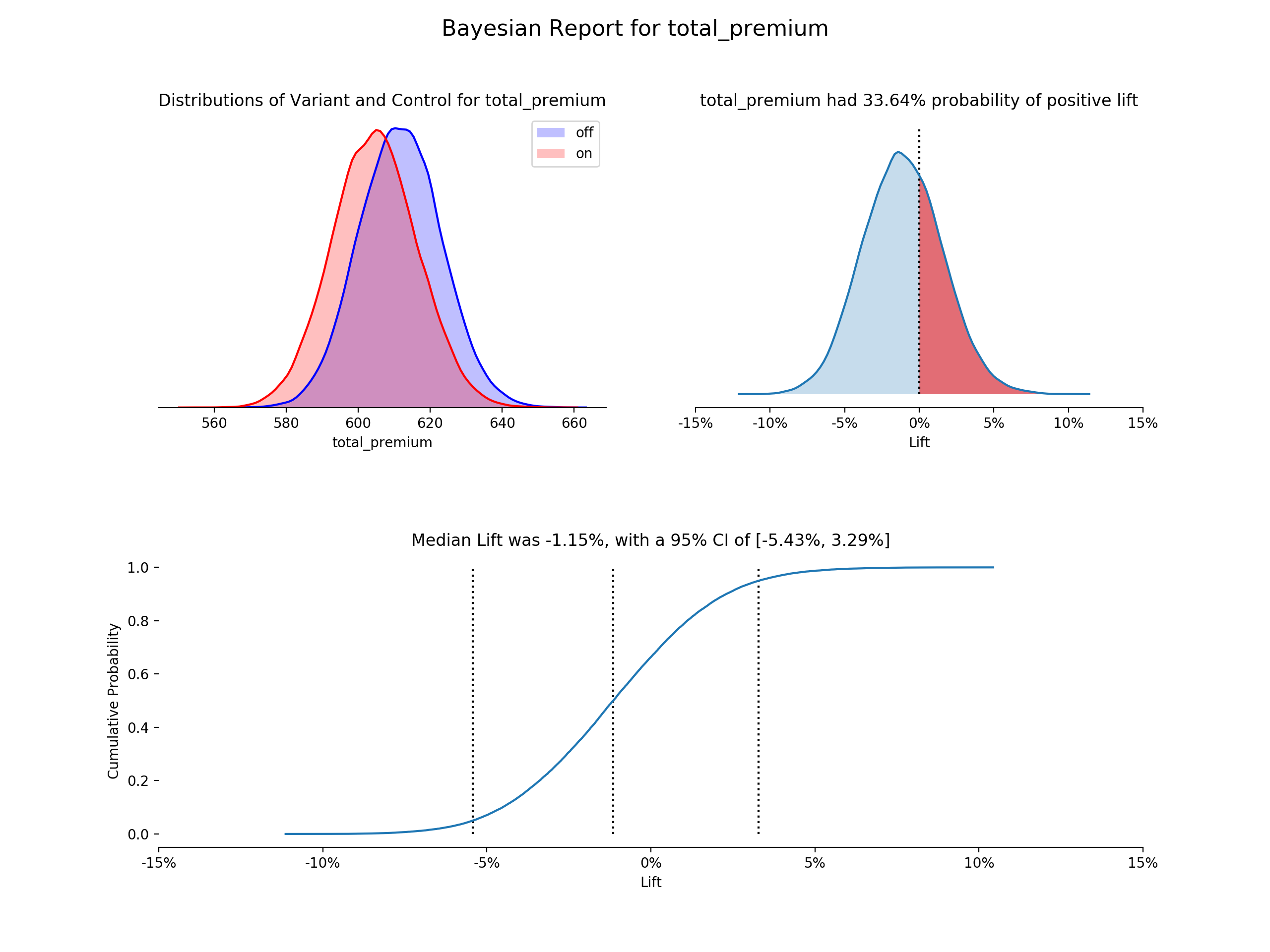
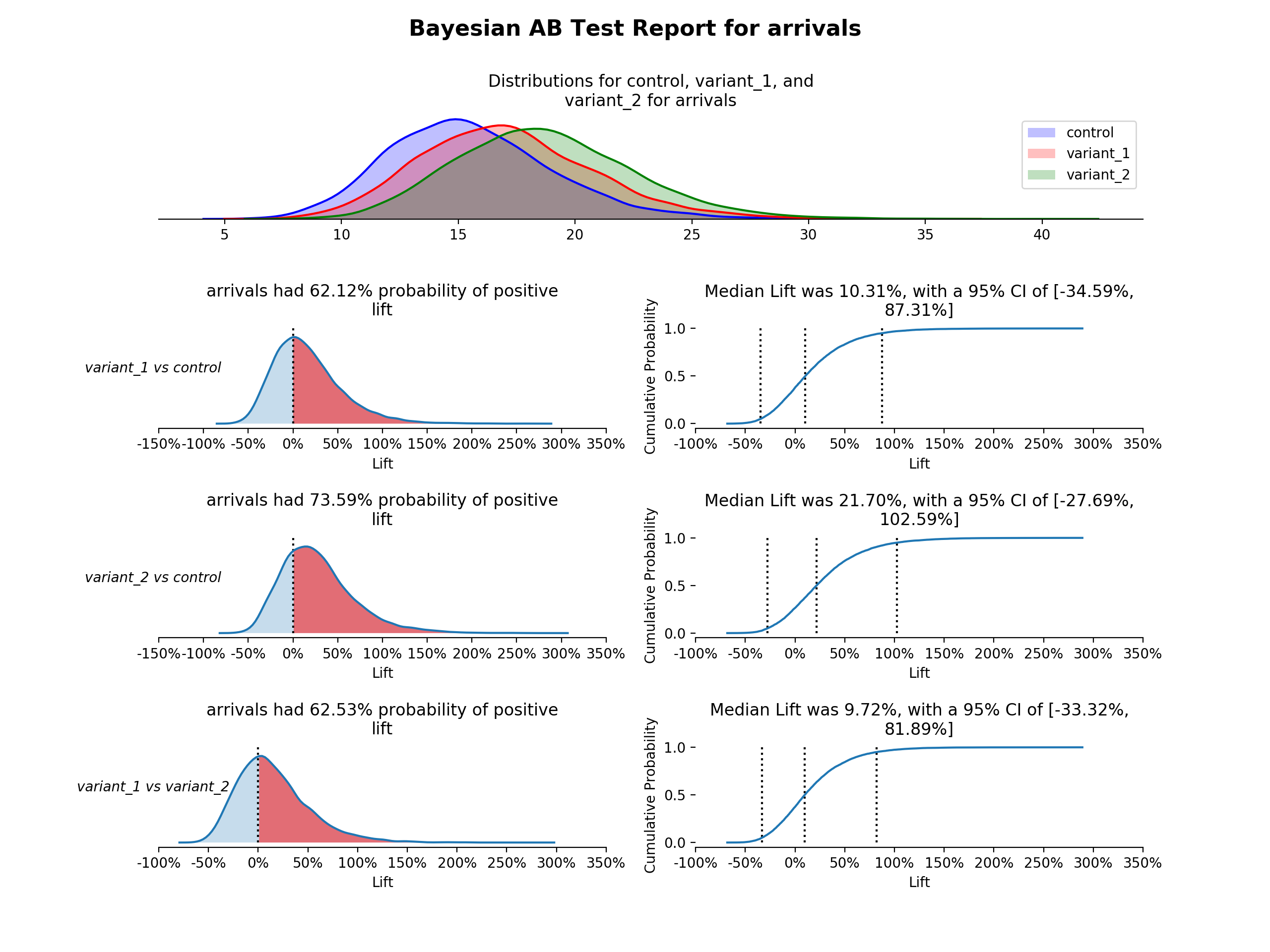
first_test.plot()- Posterior Distributions - Shows both buckets posterior distributions, an estimate of the true conversion rate or continuous value, based on the prior and the observed data (using kernel density estimation plots)
- Lift PDF - Shows the probability density distribution of lift for the variant over the control, an estimate of the chance of improvement of the variant over the control (using kernel density estimation plot). Calculated by: the results from the sampled values for the variant percent change from the control. The proportion greater than 0 is highlighted.
- Empirical CDF of Liftå - Uses the same lift vector, but is the empirical cumulative distribution function. Median lift is highlighted and labeled, as well as the chosen confidence interval.
0.1.0- 12/02/20191.0.0- 12/27/20191.0.1- 01/02/20201.0.2- 02/11/20201.0.3- 02/21/2020
There is a lot of documentation out there about a Bayesian framework of A/B testing. Some of the specific articles are listed below. Most of the work I came across was written in R, and I set out to create a Python implementation. The visuals were inspired by a standard template we use at Root, first written by https://github.com/zachurchill-root.
Articles Reference: