This is another real-time scaffolder beside npScarf. Instead of using contig sequences as pre-assemblies, this tool is able to work on assembly graph (from SPAdes). The batch algorithm has been implemented in hybrid assembler module of Unicycler and others.
npScarf is the real-time hybrid assembler that use the stream of long reads to bridge the Illumina contigs together, expecting to give more complete genome sequences while the sequencing process is still ongoing. The pipeline has been applied sucessfully for microbial genomics and even bigger data sets. However, due to its greedy approach over the noisy data, it is difficult to eliminate all mis-assemblies without further pre-processing and parameter tuning. To help prevent this issue, the assembly graph - bulding block graph structure for the contigs - should be used as the source for bridging algorithm. This approach can give better accuracy, but as the trade-off, are more computational expensive and challenging to adapt in the real-time mode.
A (rather simple at the moment) Graphical User Interface is implemented for better interaction. GraphStream, a dynamic graph library for Java, has been employed for such task. At the moment, only a few among sea of amazing features of this library had been used, leaving many room for visualiazation improvements.
For detail options of the commandline interface:
java -cp target/assembly-x.x.x-SNAPSHOT.jar org.rtassembly.NPGraphCmd -h
Usage:
Options:
--si=s Name of the short-read assembly file.
(default='')
--sf=s Format of the assembly input file. Accepted format are FASTG, GFA
(default='')
--li=s Name of the long-read data input file, - for stdin.
(default='')
--lf=s Format of the long-read data input file. This may be FASTQ/FASTA (MinION reads) or SAM/BAM (aligned with the assembly graph already)
(default='')
--output=s Output folder for temporary files and the final assembly npgraph_assembly.fasta
(default='/tmp/')
--sb=s Name of the metaBAT file for binning information (experimental).
(default='')
--aligner=s Aligner tool that will be used, either minimap2 or bwa
(default='')
--algOpt=s Settings used by aligner to align long reads to the contigs
(default='')
--msa=s MSA tools for consensus. Options include spoa, kalign3 (fast); kalign2, poa (slow).
(default='')
--overwrite Whether to overwrite or reuse the intermediate file
(default='true')
--sp Whether to use SPAdes contigs.paths for bridging.
(default='false')
--qual=i Minimum quality of alignment to considered
(default='10')
--mcov=i Minimum number of reads spanning a confident bridge
(default='3')
--depth=i Maximum depth for searching path between 2 neighbors
(default='300')
--anchor=i Minimum length for being considered as an anchor contig.
(default='1000')
--unique=i Minimum length for being assumed a unique contig.
(default='10000')
--gui Whether using GUI or not.
(default='false')
--keep Whether to keep extremely-low-coveraged contigs.
(default='false')
--verbose For debugging.
(default='false')
--help Display this usage and exit
(default='false')
or invoke the GUI of the module and control it interactively by
java -cp target/assembly-x.x.x-SNAPSHOT.jar org.rtassembly.NPGraphCmd -gui
A proper combination of command line and GUI can provide an useful streaming pipeline that copes well with MinION output data. This practice allows the assembly to take place abreast of nanopore sequencing run.
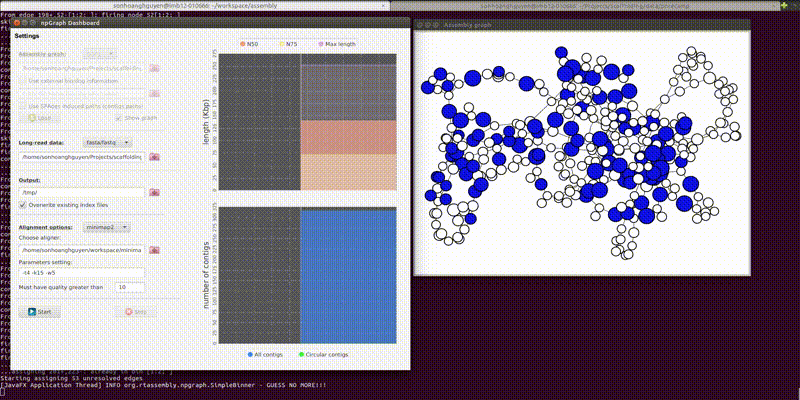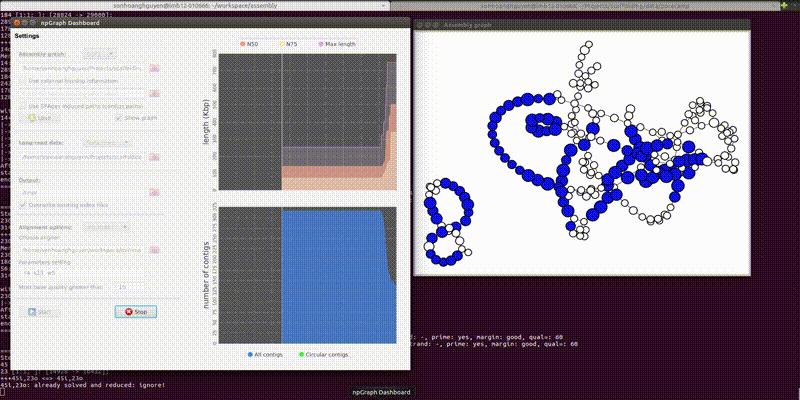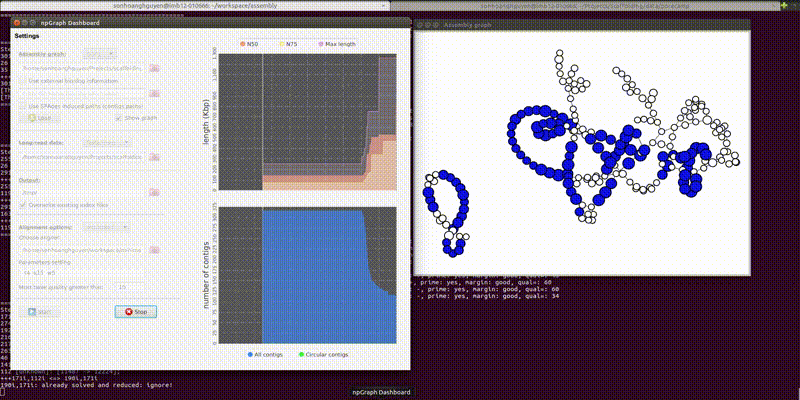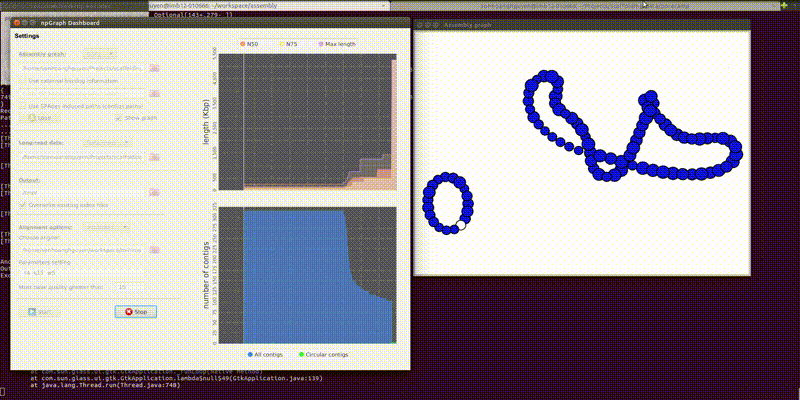
The GUI includes the dashboard for control the settings of the program and another pop-up window for a simple visualization of the assembly graph in real-time.
In this mode, I purposely separate the assembly graph loading stage from the actual assembly process, so user need to have a legal (and decent) graph first before carry out any further tasks. After that, user can choose to integrate an aligner (bwa or minimap2) to npGraph to invoke from it, or run the alignment independently and provide SAM/BAM as input to npGraph for the next stage of bridging and assembly.
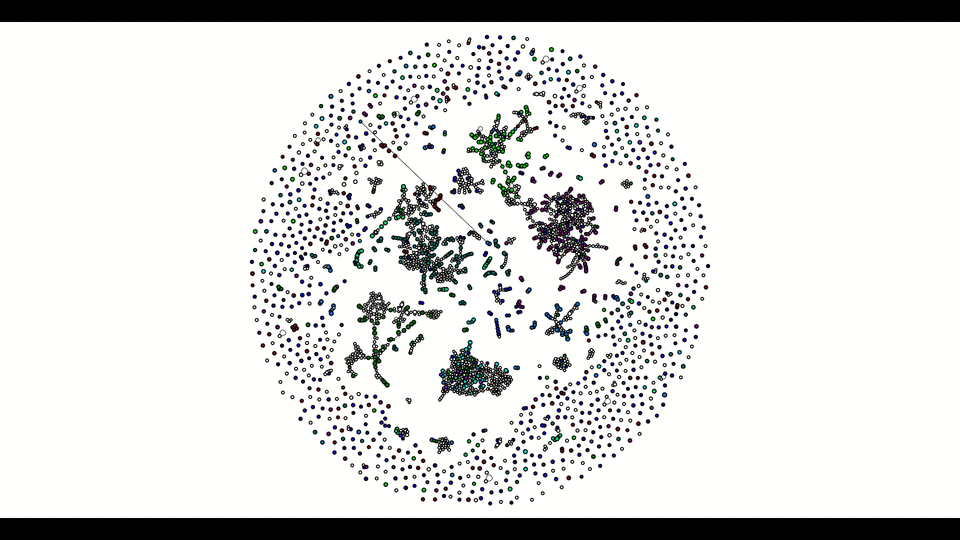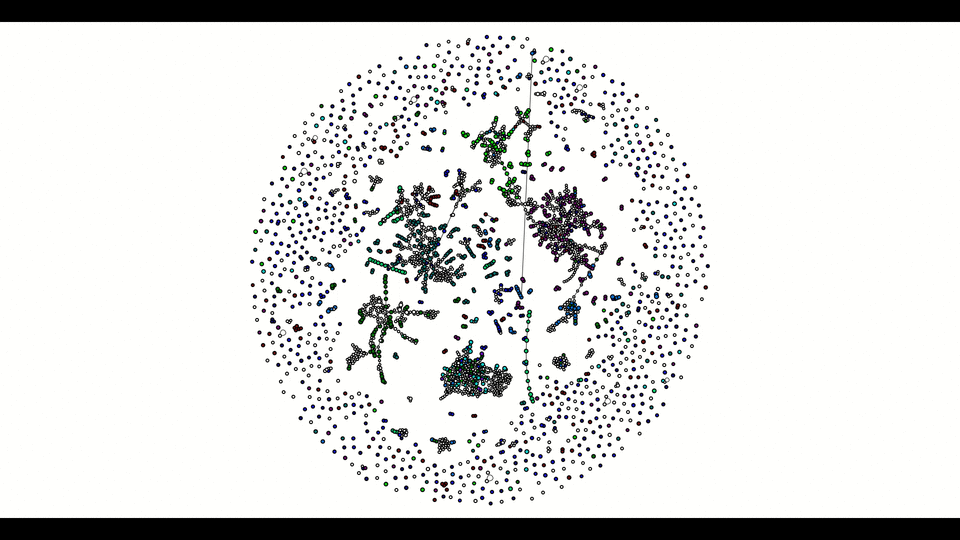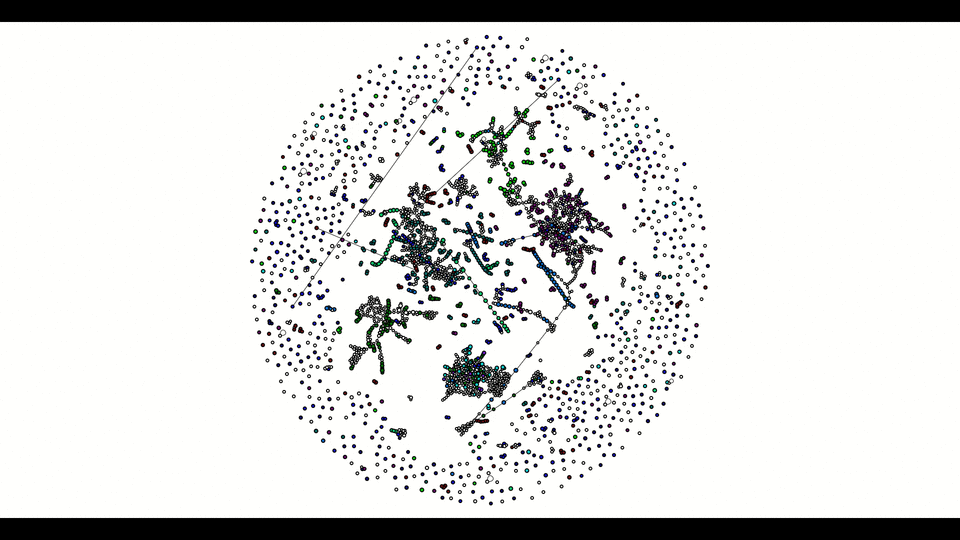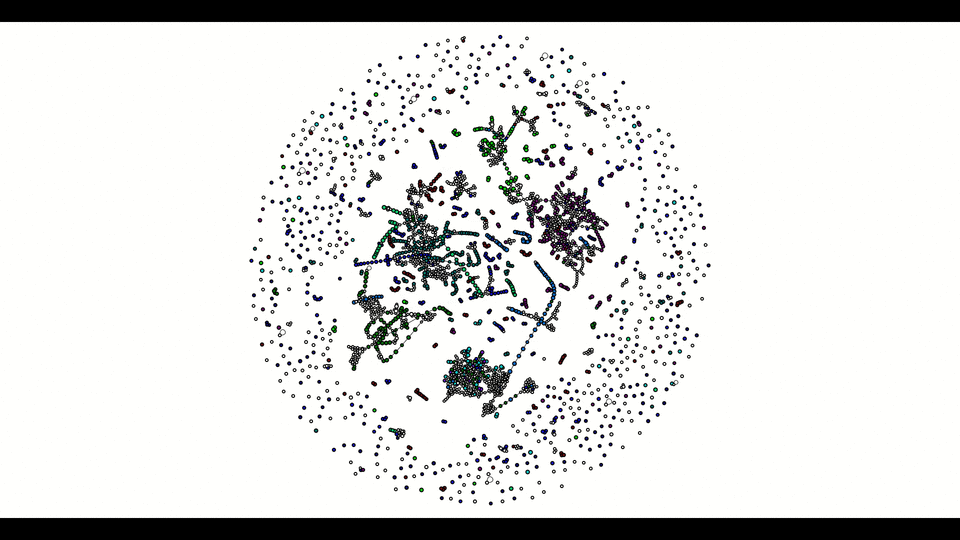
From the second window, the colored vertices are unique contigs while the white ones are either unknown or repeats. The number of colors (other than white) indicates number of populations (e.g. chromosome vs plasmids, or different bins in metagenomics). Any edge represendted is bi-directed, the final result will only include edges following the flow properly. The STOP button can prematurely terminate the assembly process and output whatever it has till that moment.
More features would be added later to the GUI but it's not the focus of this project. I will output GFA files representing graph in different timestamps if users want to investigate more the details of the graph structure later (e.g. by using Bandage).
All settings from the GUI can be set beforehand via commandline interface. Without using GUI, the mandatory inputs are assembly graph file (-si) and long-read data (-li). The assembly graph must be output from SPAdes in either FASTG or GFA format (normally assembly_graph.fastg or assembly_graph.gfa). From new version of SPAdes, the output GFA file is assembly_graph_with_scaffolds.gfa which includes SPAdes path finding and scaffolding results. Sometimes, this might give additional mis-assemblies so the original graph of the building-block contigs is preferred.
The long-read data will be used for bridging and can be given as DNA sequences (FASTA/FASTQ format, possible .gz) or alignment records (SAM/BAM) as mentioned above. npGraph will try to guess the format of the inputs based on the extensions, but sometimes you'll have to specify it yourself (e.g. when "-" is provided to read from stdin). If the sequences are given, then it's mandatory to have either minimap2 (recommended) or BWA-MEM installed in your system to do the alignment between long reads and the pre-assemblies.
Alternative option is to use your favourite aligner and provide SAM/BAM to npGraph. In this case, you have to manually convert FASTG/GFA file to FASTA file to use as the reference for the alignment. You can use awk script to do this, e.g. for converting FASTG file
awk -F '[:;]' -v q=\' 'BEGIN{flag=0;}/^>/{if(index($1,q)!=0) flag=0; else flag=1;}{if(flag==1) print $1;}' assembly_graph.fastg > assembly_graph.fasta
or if the graph file is GFA v1 we can use
awk '/^S/{print ">"$2; print $3;}' assembly_graph.gfa | fold > assembly_graph.fasta
Note that GFA format from SPAdes is preferred over FASTG since the former gives hint about the k-mer parameter and others, also it is becoming the standard for assembly graph that adapted by many other software. And then you can generate SAM/BAM file with our recommended parameters:
minimap2 -t16 -k15 -w5 -a assembly_graph.fasta nnp.fastq ...
or
bwa mem -t16 -k11 -W20 -r10 -A1 -B1 -O1 -E1 -L0 -a -Y assembly_graph.fasta nnp.fastq ...
It is important to emphasis the quality of the assembly graph to the final results. Unicycler pre-process the graph data by running SPAdes with multiple kmer options to chose the best one. npGraph v0.2 will use the best graph suggested by SPAdes when running through -k 55,77,99,107,127 (normally at k=127 for 2x150 Illumina pair-ended reads) and then try to scan the contigs' endings to find potential shorter overlaps and fix the dead-ends. Choosing the right kmer for your data is important but npgraph favours the big values over small ones (k<99).
At the moment, npGraph input is a single graph file given in FASTG or GFA, includes but not limited to SPAdes output. The plan of iterating over several graph files generated by the same short-read data (of different setttings and/or assemblers) to chose the best one is considered for latter release.
Normally, 60X Illumina MiSeq data would give decent SPAdes assembly graph. The better the assembly graph is, the more complete and accurate assembly you'll get. It doesn't do neither any polishing or other exhaustive post-processing for the final assembly assuming the quality is equivalent to the short-read data which is decent enough.
Beside the default binning algorithm, npgraph is trying to use binned results from MetaBAT for more complicated graph, such as metagenomics. The bin file is a text file that present <contig_ID> <bin_ID> in each line. Below is an example to generate one.
The tool generates the assembly in a FASTA file npgraph_assembly.fasta and a GFA v1 file npgraph_assembly.gfa . The FASTA file output contains the significant contigs only while the GFA file contains all the original contigs and the links between them. The repetitive elements will be duplicated in the GFA output for easier examination. For example, we can see from the second column of the S(egment) line somethings like <X.Y> means this is the Yth copy of the original contig X (without suffix). For that reason, we can see how many copies for each contigs in the final result by
awk '/^S/{print}' npgraph_assembly.gfa|sort -nk2,2|less -S
Also, the stats of the assembly process is reported in the GUI dashboard and/or stdout as the program is running. Note that the ultimate output graph (when you hit Stop button from the dashboard or when the EOF of the long read data is reached) is undergone a post-processing step to greedily connect insignificant bridges.
npGraph can be applied to assemble genome of complex isolates or metagenomics. Depend on the complexity of the data set, additional pre-processing should be considered. Below is an example of the fundamental steps for a metagenomics assembly.
#1. run metaSPAdes to have the assembly
~/sw/spades/current/bin/spades.py --meta -k 55,77,99,107,127 --pe1-1 nextSeq_1_PE.fastq --pe1-2 nextSeq_2_PE.fastq -o metaSPAdes
#2. convert assembly graph to a FASTA file
awk -F'[:;]' -v q="'" '/^>/{if(index($1,q) ==0 ) flag=1; else flag=0;} {if(flag) print $1}' metaSPAdes/assembly_graph.fastg > metaSPAdes/assembly_graph.fasta
#3. alignt pair-end reads to the assembly graph contigs + sort the bam file
~/sw/bwa.kit/bwa index metaSPAdes/assembly_graph.fasta
~/sw/bwa.kit/bwa mem -t8 metaSPAdes/assembly_graph.fasta nextSeq_1_PE.fastq nextSeq_2_PE.fastq | ~/sw/samtools-1.4/samtools sort -@8 -o reads2assembly.bam -
#4. run metaBAT with --saveCls --noBinOut to have the binning information
~/sw/metabat/jgi_summarize_bam_contig_depths --outputDepth depth.txt reads2assembly.bam
~/sw/metabat/metabat2 --saveCls --noBinOut --inFile metaSPAdes/assembly_graph.fasta --abdFile depth.txt --outFile metabat/bin
Below is how it looked like using npGraph with a mock community of 11 species from PoreCamp.
- GUI consumes quite a lot memory, considering increase JVM heap size (java -Xmx) if it's slow to response. If the number of vertices in the assembly graph greater than 10000, you shouldn't show the graph in real-time if running on a normal desktop.
- If aligner is used along side, there is more resource occupied. Considering separate alignment and npGraph+GUI on different machines communicating through network socket e.g. by Japsa utility jsa.util.streamServer and jsa.util.streamClient
- Most suitable for bacterial data (assembly graph not too complicated). Consider using npScarf for bigger data set.
- To keep the lightweight property that suitable for real-time analysis, a quick alignment-based voting & scoring system is implemented for the path finding over the candidate nodes instead of the expensive consensus phasing. minimap2 is also recommended over bwa due to its speed, however sometimes bwa returns more sensitive alignments than the former.
Publication on the way.