Automatized phylogenetic tree building from bacterial core genomes.
An article describing bcgTree is published in Genome.
If you use bcgTree for your research please cite:
Also please cite the external programs and the source of the HMMs (see Background section) if you use the default essential.hmm
If you like the tool consider voting for it on LabWorm.
See this file for instructions on how to reproduce results from our article.
Please note that some of the dependencies have their own Licenses.
To execute bcgTree perl5 is required along with the following modules (should be part of the core installation, but also available via cpan):
- Getopt::Long
- Pod::Usage
- FindBin
- File::Path
- File::Spec
The following modules are included in this repo:
- Log::Log4perl (Artistic License)
- Getopt::ArgvFile (Artistic License)
- perl5lib-Fasta (MIT License)
- perl5lib-Fastq (MIT License)
- perl5lib-Verbose (MIT License)
To use the graphical user interface a Java Runtime Environment (JRE, tested with version 8) is required. The following extra frameworks are used (included):
bcgTree is a wrapper around multiple existing tools. The following external programs are called by bcgTree and have to be installed. The specified versions are the ones we used for testing (older versions might or might not work). Newer versions should work (otherwise feel free to open an issue).
- hmmsearch (HMMER version 3.1b1) - Eddy et al, 2010. HMMER3: a new generation of sequence homology search software.
- muscle (v3.8.31) - Edgar et al, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
- Gblocks (version 0.91b) - Castresana et al, 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552.
- RAxML (version 8.2.4) - Stamatakis et al, 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Additionally SeqFilter (Hackl et al, 2014. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004-3011) is needed but it is included in this repo.
There are two hmm files included in the data directory. Both contain HMMs from TIGRFAM (LGPL) and PFAM (CC0). The ubcg.hmm file is reproduced from UBCG as described in Na, SI., Kim, Y.O., Yoon, SH. et al. J Microbiol. (2018) 56: 280.
You can download the latest release from here. After extraction you can start the graphical interface by double clicking the bcgTree.jar file in the bcgTreeGUI folder. Please remember to install the required external programms (see above). Alternatively you can start the perl script bcgTree.pl in the bin folder from the command line. Using the command line interface is the recommended way. You can also clone the bcgTree git repository by executing the following commands:
git clone https://github.com/molbiodiv/bcgTree.git
# Now you can run bcgTree.pl
bcgTree/bin/bcgTree.pl --help
# Or start the java GUI
cd bcgTree/bcgTreeGUI
java -jar bcgTree.jarThe graphical user interface is just a convenient way to call bcgTree. Usage is meant to be self-explanatory, any suggestions for improvement are welcome (please open an issue). In fact it adds no extra functionality it just collects the parameters and calls the perl script on the command line. The output is written to a text field as well as a log file (bcgTree.log in the output folder). In order to preserve replicability all parameters are written in a file called options.txt in the output folder. The internal call of bcgTree is then just:
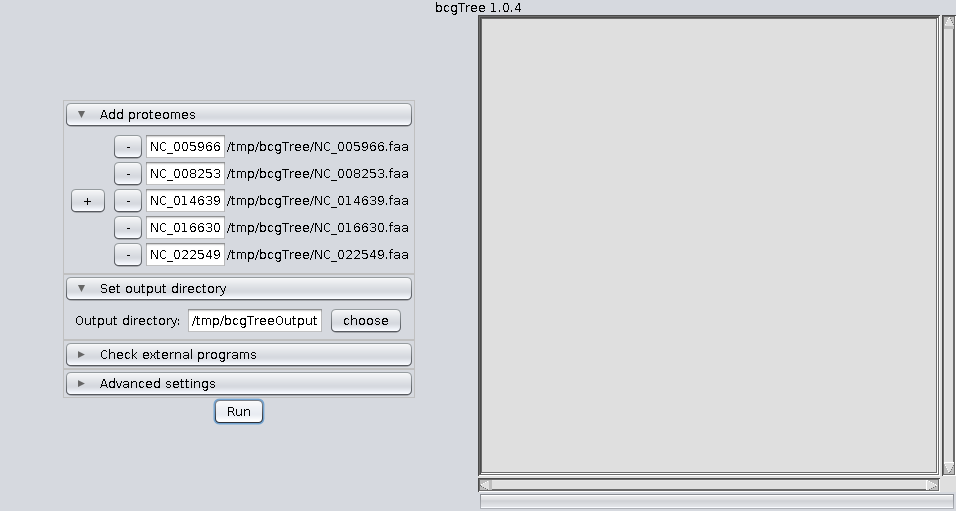
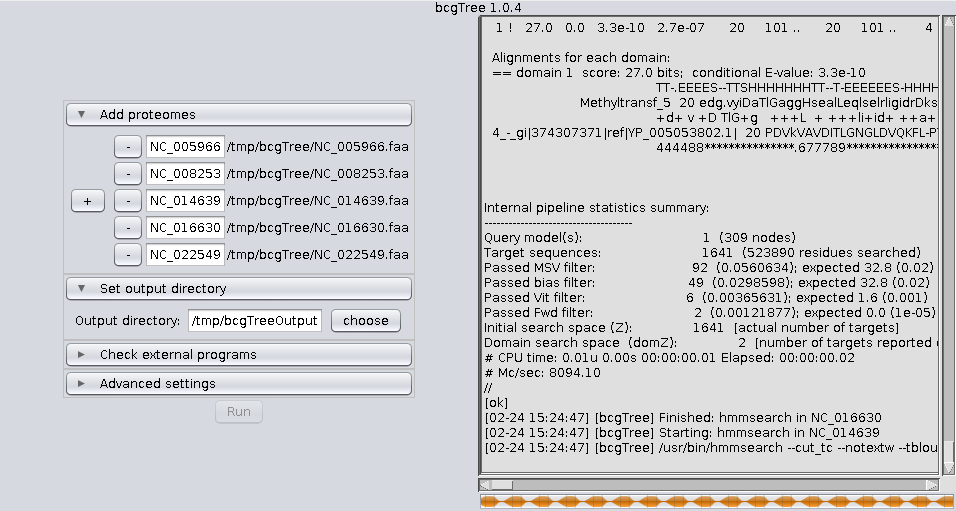
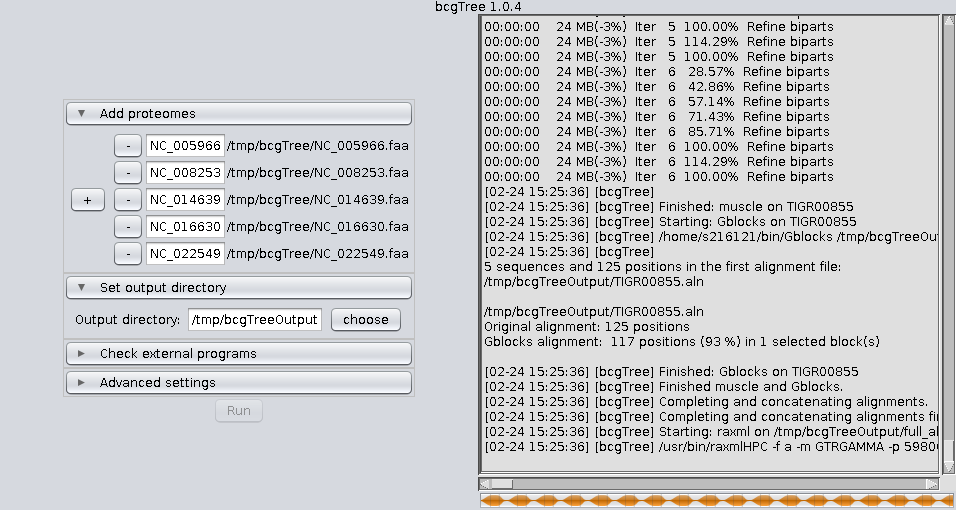
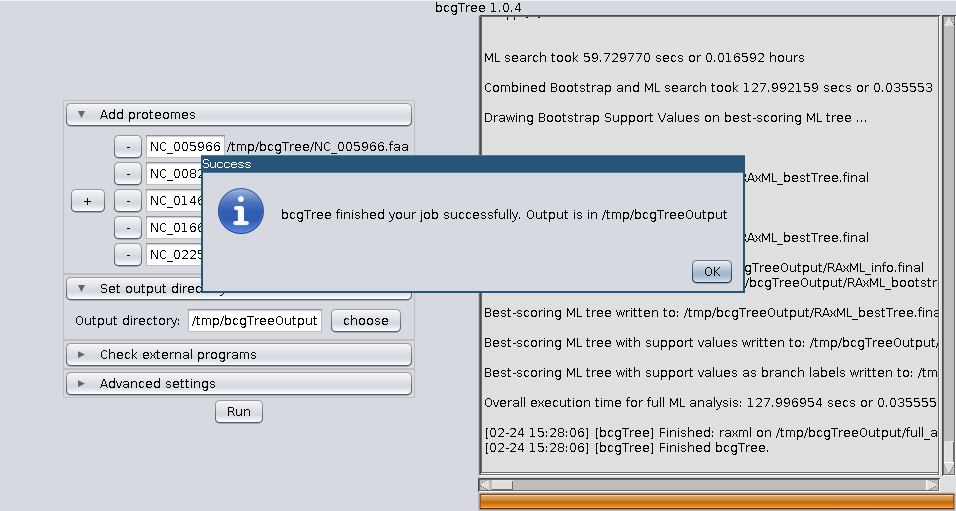
bcgTree.pl @outdir/options.txtIf you feel comfortable using the command line, calling the perl script directly is still the recommendet way to use bcgTree. Nevertheless here are some screenshots of the GUI:
Usage:
$ bcgTree.pl [@configfile] --proteome bac1=bacterium1.pep.fa --proteome bac2=bacterium2.faa [options]
Options:
[@configfile] Optional path to a configfile with @ as prefix.
Config files consist of command line parameters
and arguments just as passed on the command
line. Space and comment lines are allowed (and
ignored). Spreading over multiple lines is
supported.
--proteome <ORGANISM>=<FASTA> [--proteome <ORGANISM>=<FASTA> ..]
Multiple pairs of organism and proteomes as
fasta file paths
[--outdir <STRING>] output directory for the generated output files
(default: bcgTree)
[--help] show help
[--version] show version number of bcgTree and exit
[--check-external-programs]
Check if all of the required external programs
can be found and are executable, then exit.
Report table with program, status (ok or
!fail!) and path. If all external programs are
found exit code is 0 otherwise 1. Note that
this parameter does not check that the paths
belong to the actual programs, it only checks
that the given locations are executable files.
[--hmmsearch-bin=<FILE>] Path to hmmsearch binary file. Default tries if
hmmsearch is in PATH;
[--muscle-bin=<FILE>] Path to muscle binary file. Default tries if
muscle is in PATH;
[--gblocks-bin=<FILE>] Path to the Gblocks binary file. Default tries
if Gblocks is in PATH;
[--raxml-bin=<FILE>] Path to the raxml binary file. Default tries if
raxmlHPC is in PATH;
[--threads=<INT>]
Number of threads to be used (currently only relevant for raxml).
Default: 2 From the raxml man page: PTHREADS VERSION ONLY! Specify
the number of threads you want to run. Make sure to set "-T" to at
most the number of CPUs you have on your machine, otherwise, there
will be a huge performance decrease!
[--bootstraps=<INT>]
Number of bootstraps to be used (passed to raxml). Default: 100
[--min-proteomes=<INT>]
Minimum number of proteomes in which a gene must occur in order to
be kept. Default: 2 All genes with less hits are discarded prior to
the alignment step. This option is ignored if --all-proteomes is
set.
[--all-proteomes]
Sets --min-proteomes to the total number of proteomes supplied.
Default: not set All genes that do not hit all of the proteomes are
discarded prior to the alignment step. If set --min-proteomes is
ignored.
[--hmmfile=<PATH>]
Path to HMM file to be used for hmmsearch. Default:
<bcgTreeDir>/data/essential.hmm
[--raxml-x-rapidBootstrapRandomNumberSeed=<INT>]
Random number seed for raxml (passed through as -x option to raxml).
Default: Random number in range 1..1000000 (see raxml command in log
file to find out the actual value). Note: you can abbreviate options
(as long as they stay unique) so --raxml-x=12345 is equivalent to
--raxml-x-rapidBootstrapRandomNumberSeed=12345
[--raxml-p-parsimonyRandomSeed=<INT>]
Random number seed for raxml (passed through as -p option to raxml).
Default: Random number in range 1..1000000 (see raxml command in log
file to find out the actual value). Note: you can abbreviate options
(as long as they stay unique) so --raxml-p=12345 is equivalent to
--raxml-p-parsimonyRandomSeed=12345
[--raxml-aa-substitiution-model "<MODEL>"]
The aminoacid substitution model used for the partitions by RAxML.
Valid options for RAxML 8.x are: DAYHOFF, DCMUT, JTT, MTREV, WAG,
RTREV, CPREV, VT, BLOSUM62, MTMAM, LG, MTART, MTZOA, PMB, HIVB,
HIVW, JTTDCMUT, FLU, STMTREV, DUMMY, DUMMY2, AUTO, LG4M, LG4X,
PROT_FILE, GTR_UNLINKED, GTR bcgTree will not check whether the
provided option is valid but rather pass it to RAxML literally.
Default: AUTO
[--raxml-args "<ARGS>"]
Arbitrary options to pass through to RAxML. The ARGS part should be
in quotes and is appended to the RAxML command as given.
The results all end up in the directory specified via –outdir (or bcgTree if none is specified). This folder contains lots of intermediate files from all steps. If the run was successful the most interesting files will be the RAxML files:
- <outdir>/RAxML_bestTree.final
- <outdir>/RAxML_bipartitionsBranchLabels.final
- <outdir>/RAxML_bipartitions.final
- <outdir>/RAxML_bootstrap.final
- <outdir>/RAxML_info.final
Further the log file (<outdir>/bcgTree.log) contains all executed commands and their output. This is useful as a reference, for re-executing steps manually and for debugging in case something went wrong. All other files are the outputs of different steps of the pipeline. Their names should be self-explanatory.
107 essential genes as described in: Dupont CL, Rusch DB, Yooseph S, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. The ISME Journal. 2012;6(6):1186-1199. doi:10.1038/ismej.2011.189. Supplementary Table S1 (which is actually an image) contains a list of the used genes and HMMs with cut-offs.
From the manuscript: “Genome completeness estimates Using the Comprehensive Microbial Resource as a database, 107 hidden Markov models (HMMs) that hit only one gene in greater than 95% of bacterial genomes were identified (Supplementary Table S1). Trusted cutoff scores for the TIGRFAMs and Pfam HMMs were those supplied by the TIGRFAMs and Pfam libraries (Haft et al., 2003; Finn et al., 2010).”
In the publication: M Albertsen, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW and Nielsen PH, Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nature Biotechnology 31, 533–538 (2013) doi:10.1038/nbt.2579 the authors use the same list of 107 genes (111 HMMs, glyS, pheT, proS and rpoC have two HMMs each) as above and provide a readily created hmm file via GitHub. This file has been used as a starting point but an error had to be fixed.
The logo has been designed by Markus J. Ankenbrand and Alexander Keller. Cliparts from openclipart.org have been used:
The font is from fontlibrary.org:
These are some tools with similar goals and approaches. If you know another one, please open a pull request to add it to the list.
- PO_2_MLSA - by @jvollme - Enables the calculation of phylogenies based on single copy core genome gene products, based on bidirectional BLAST results obtained with proteinortho.
- UBCG - by Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I. & Chun, J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56. Paper
- Add parameter
--genomewith translation viaprodigal(#15) - Update and improve documentation (#19, #35, #38, #44)
- Make GUI independent of working directory (#33)
- Breaking: the default aa substitution model for RAxML changed from WAG to AUTO.
This has an impact on performance (it is faster to set this parameter to a fixed value).
To get the same behaviour as in earlier versions pass
--raxml-aa-substitution-model=WAG - Add parameter
--raxml-aa-substitution-model(#29) - Add HMMs of UBCG (#25)
- Fix GUI, add scrollbar (#23)
- Add parameter –raxml-args (#22)
- Add parameters –min-proteomes and –all-proteomes (#21)
- Set default bootstraps to 100
- Add description for reproduction of results in paper
- Add logo to GUI
- Improve layout (avoid errors with large text fields)
- Update jar file
- Add advanced settings and external programs to GUI
- Add GUI screenshots to README
- Finish GUI layout
- Fix outdir bug (manually entered text was ignored)
- Update documentation in README
- Improve layout of GUI (proteomes panel)
- Add parameter to check external programs
- Fix SeqFilter dependencies
- Add swingx and own accordion element for GUI
- Improve GUI design (GridBagLayout)
- Add log4perl and Getopt::ArgvFile to package (simplify installation)
- Remove Bioperl dependency
- Add submodules directly (SeqFilter)
- Update documentation
- Add java GUI