| Branch |  |
 |
 |
|---|---|---|---|
master |
|||
develop |
R package for TMHMM [1, 2], to predict transmembrane helices in proteins.
Please note that this page is intended for academic users only. Other users are requested to contact the Software Package Manager at [email protected]
usethis::install_github("richelbilderbeek/tmhmm")
Install TMHMM to a default folder:
library(tmhmm)
install_tmhmm("https://services.healthtech.dtu.dk/download/28c408dc-ef5e-47ad-a284-66754bcd27f7")
The URL can be obtained by requesting a download link at
the TMHMM website
at https://services.healthtech.dtu.dk/service.php?TMHMM-2.0.
As this URL expires after four hours, tmhmm cannot do this for you.
library(tmhmm)
# Use an example proteome
fasta_filename <- system.file(
"extdata",
"UP000464024.fasta",
package = "tmhmm"
)
# Predict the topology
topology <- predict_topology(fasta_filename)
# Simplify the protein names
topology$name <- stringr::str_match(
string = topology$name,
pattern = "..\\|.*\\|(.*)_SARS2"
)[,2]
# Plot the topology
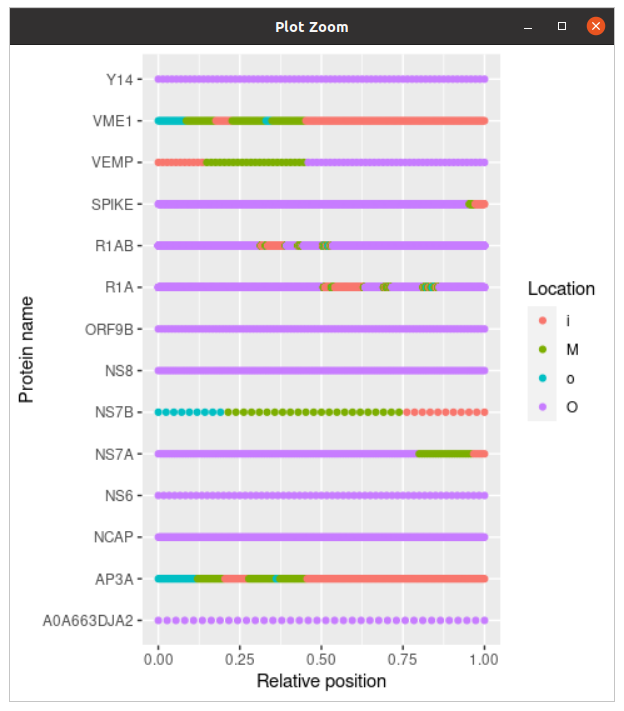
plot_topology(topology)Each element of the topology sequence denotes the location of each amino acid:
i: insideo: outsideM: membrane
tmhmm can only work on the set of operating systems TMHMM
works on. Currently, only Linux is supported:
| Operating system | Supported by TMHMM | Supported by tmhmm |
|---|---|---|
| AIX | Y | ? |
| IRIX32 | Y | ? |
| IRIX64 | Y | ? |
| Linux | Y | Y |
| MacOS | N | N |
| OSF1 | Y | ? |
| SunOS | Y | ? |
| Windows | N | N |
These are the ones I use:
- pureseqtmr:
predict membrane protein topology at two levels (TMH or non-TMH),
FOSS (i.e. no registration forms needed, thus easy install)
Note that running script shows that TMHMM is approximately 100x faster than PureseqTM:
fasta_filename <- system.file("extdata", "UP000464024.fasta", package = "pureseqtmr")
system.time({tmhmm::predict_topology(fasta_filename = fasta_filename)})
system.time({pureseqtmr::predict_topology(fasta_filename = fasta_filename)})
Results:
> system.time({tmhmm::predict_topology(fasta_filename = fasta_filename)})
user system elapsed
1.723 0.008 1.733
> system.time({pureseqtmr::predict_topology(fasta_filename = fasta_filename)})
user system elapsed
117.511 1.746 85.748
Also when running these tools from scripts, the results are similar:
time ./bin/decodeanhmm.Linux_x86_64 lib/TMHMM2.0.model UP000464024.fasta -f lib/TMHMM2.0.options
[...]
real 0m1.379s
user 0m1.375s
sys 0m0.004s
time ./PureseqTM_proteome.sh -i UP000464024.fasta
71
real 0m53.471s
user 1m36.354s
sys 0m1.426s
-
[1] A. Krogh, B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305(3):567-580, January 2001.
-
[2] E. L.L. Sonnhammer, G. von Heijne, and A. Krogh. A hidden Markov model for predicting transmembrane helices in protein sequences. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen, editors, Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, pages 175-182, Menlo Park, CA, 1998. AAAI Press.