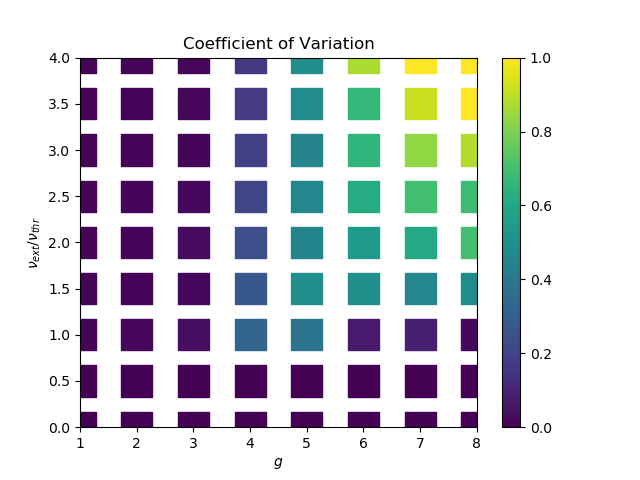
A short introduction to snakemake based on a parameter scan of the Brunel network. Final result:
- understand the
Snakefile- compare to the visualization
graph.png - produce the visualization yourself:
snakemake --dag | dot | display - make a dry-run:
snakemake -np - understand the interplay of the
Snakefileandconfig.yaml
- compare to the visualization
- produce a first version of the phase diagram
- execute the workflow:
snakemake - have a look at the phase diagram
- execute the workflow:
- extend the phase diagram a bit
- extend the parameter range using
config.yaml - make a dry-run:
snakemake -np - why did nothing happen?
- force the execution:
snakemake -f plotPhaseDiagram -np - run using multiple threads:
snakemake -f plotPhaseDiagram --cores 3
- extend the parameter range using
- let's stop using a downscaled network and a rather short simulation time
- understand the
N_scaleoptions:python scripts/simulateBrunel.py --help - add an
N_scaleparameter to the Snakefile and pass it as an argument - repeat the same with the
simtime - play around a bit and see how it affects the statistics
- does the rate change with
N_scale? why / why not?
- understand the
- run a full scan of the parameter regime
- change
config.yamltoconfig_full.yaml - use multiple cores to simulate multiple parameter sets in parallel
- go grab a coffee, do something fun, or advance to the next task
- change
- let's think about a more modular implementation of
simulateBrunel.py- look at
simulateBrunelModular.pyand decide for yourself if this is cleaner - besides code structure, why is could this implementation be useful?
- look at
brunel_parameters.yaml: parameter file containing all network parametersconfig.yaml: configuration file for the snakemake worflowconfig_full.yaml: configuration file for the snakemake worflow corresponding to the above figuregraph.png: visualization of the snakemake workflowSnakefile: snakemake workflow filescripts/simulateBrunel.py: script to simulate a Brunel network (naive implementation)scripts/simulateBrunelModular.py: script to simulate a Brunel network (modular implementation)scripts/plotPhaseDiagram.py: script to plot the phase diagram of the Brunel network
- execute using default settings:
snakemake - list all rules:
snakemake -l - summarize workflow output status:
snakemake -S - dry-run mode, print bash commands:
snakemake -np - visualize workflow:
snakemake --dag | dot | display - execute a specific rule:
snakemake plotPhaseDiagram - force execution of a specific rule:
snakemake -f plotPhaseDiagram - specify config file:
snakemake --configfile config_full.yaml - use multiple threads:
snakemake --cores 3