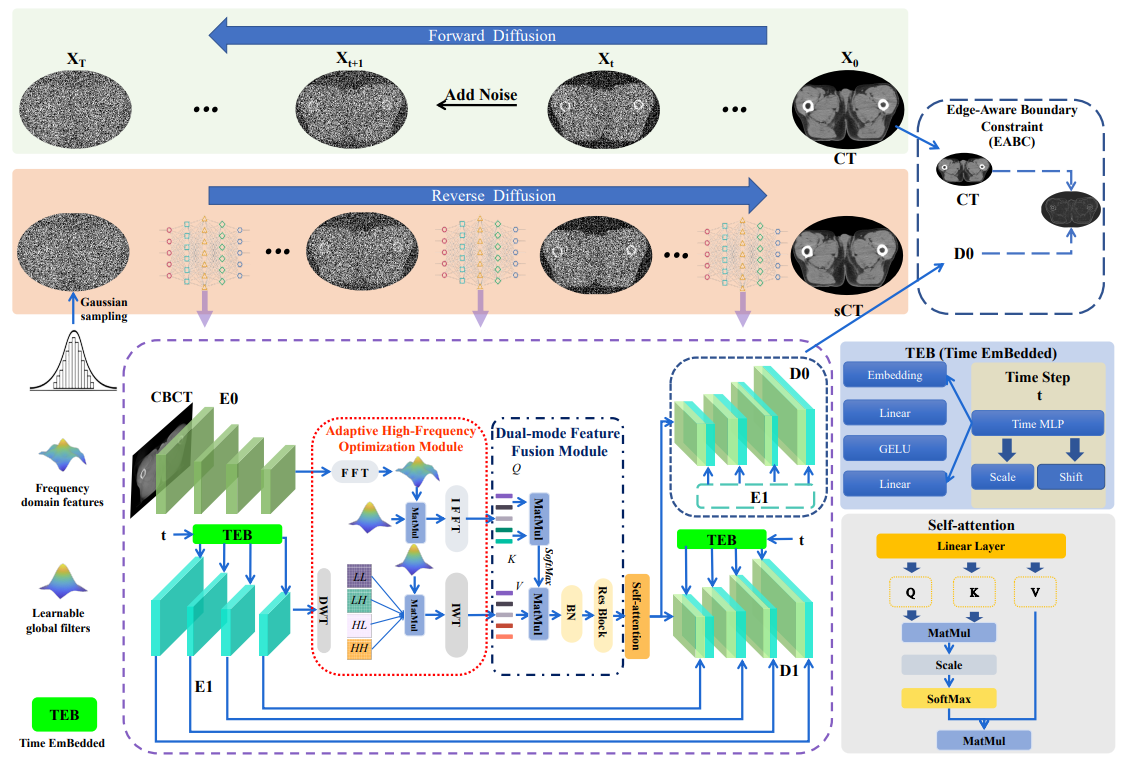
This repository implements a texture-preserving diffusion model for CBCT-to-CT synthesis, designed to improve the quality and accuracy of medical image synthesis.
data_path/
2PA092
├── CT
│ ├── 0.npy
│ ├── 1.npy
│ ├── ...
├── CBCT
│ ├── 0.npy
│ ├── 1.npy
│ ├── ...
2PA093
├── CT
│ ├── 0.npy
│ ├── 1.npy
│ ├── ...
├── CBCT
│ ├── 0.npy
│ ├── 1.npy
│ ├── ...
...
The dataset links used in this study are as follows:
Before using the dataset, convert the nii.gz data to npy format using reg/nii2npy.py, and perform registration using syn.
All the data is standardized using the mean and standard deviation of the raw data. The data processing is handled in dataloader.py
albumentations==1.3.0
einops==0.6.1
matplotlib==3.5.2
numpy==1.21.5
opencv_contrib_python==4.7.0.72
Pillow==9.5.0
scikit_image==0.19.2
scikit_learn==1.2.2
SimpleITK==2.2.1
timm==0.6.12
torch==2.0.0
tqdm==4.64.1
git clone https://github.com/zyj15416/TPDM-CBCT2CT.git
cd TPDM-CBCT2CT
python train.py, the hyperparameter settings are handled in `utils/utils.py`.
You can use `predict.py` to load the weights and test the dataset, and you can modify the number of samples to improve the inference time.
We have released pretrained weights for cbct->ct in SynthRAD2023 challenge dataset(https://pan.baidu.com/s/1ATAeiGnNeaF1G0E1z974KQ?pwd=k7m4 ). You can save these weights in relevant checkpoints folder and perform inference.
After downloading the pretrained weights, place them in the corresponding folder, such as `./weights/weights.pth`. Then, replace the weight path in `predict.py` accordingly to perform model inference.
A limited set of preprocessed CBCT and CT samples is provided in `sample_test01` to facilitate validation of the model's performance.
If you find this repository useful for your research, please use the following.
{
}