SkinSpatial is a repository for the code to analyse skin spatial data. Here we host an official version and development of STRISH (Spatial Transcriptomic and RNA In-situ Hybridization) pipeline.
STRISH is a computational pipeline that enables us to quantitatively model cell-cell interactions by automatically scanning for local expression of RNAscope data to recapitulate an interaction landscape across the whole tissue.
STRISH pipeline consists of three major steps.
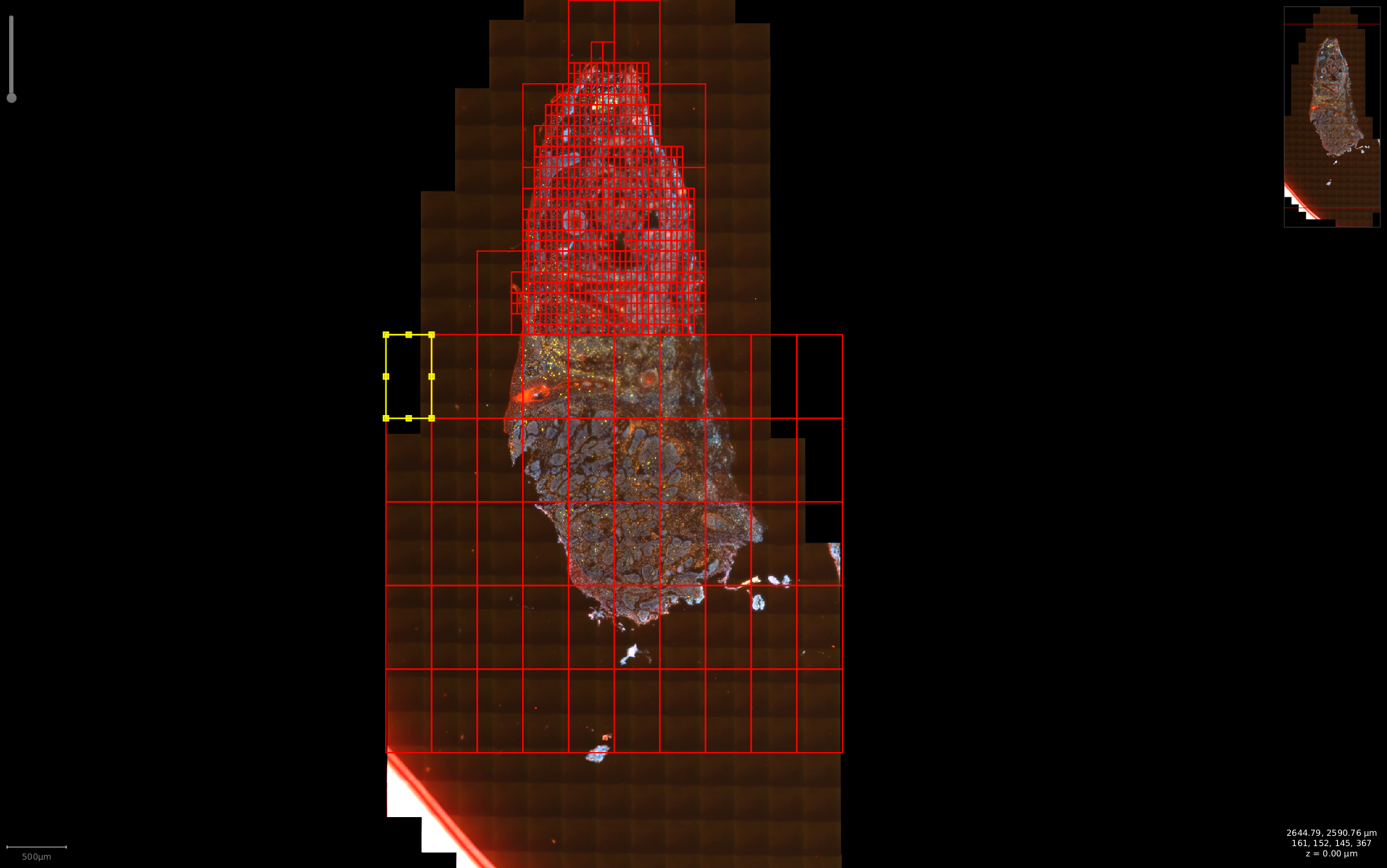
Step 1: Run cell detection for RNAScope data using series of nonoverlapped windows with QuPath. The size of scanning windows are initially set to 10% (customizable) of the whole scan image. If the cell detection result in the current window is greater than the threshold, gradually split the window size into smaller subwindow until the conditions are satisfied (window). Otherwise, remove the window from the current list of scanning windows. The cell detection algorithm pseudocode is followed pseudocode
More illustrations about STRISH cell detection with the windows strategy can be found here
Step 2: Run a Python process to computationally quantify ligand-receptor local co-expression level of two pairs of target markers. STRISH pipeline iterates all the detection results from previous step and considers every window that cells express both ligand and receptor marker. Otherwise, the coexpression score for the windows is set to 0. For the pair of ligand-receptor which two markes come from two separated scanning image, image registration is required before evaluating local co-expression level in the tissue. The final outcome is visualize by the heatmap.
Step 3: Cropping background and plotting tissue contour to focus on the main tissue area, i.e. Fig 3.
- For details about cell-cell interation analysis with Visium data, visit stLearn github page at stLearn
- For CellPhoneDB method for ligand-receptor interaction prediction, visit CellPhoneDB github page CellPhoneDB
- NicheNet analysis with Seurat object for cell-cell interaction was used following vignette: NicheNet