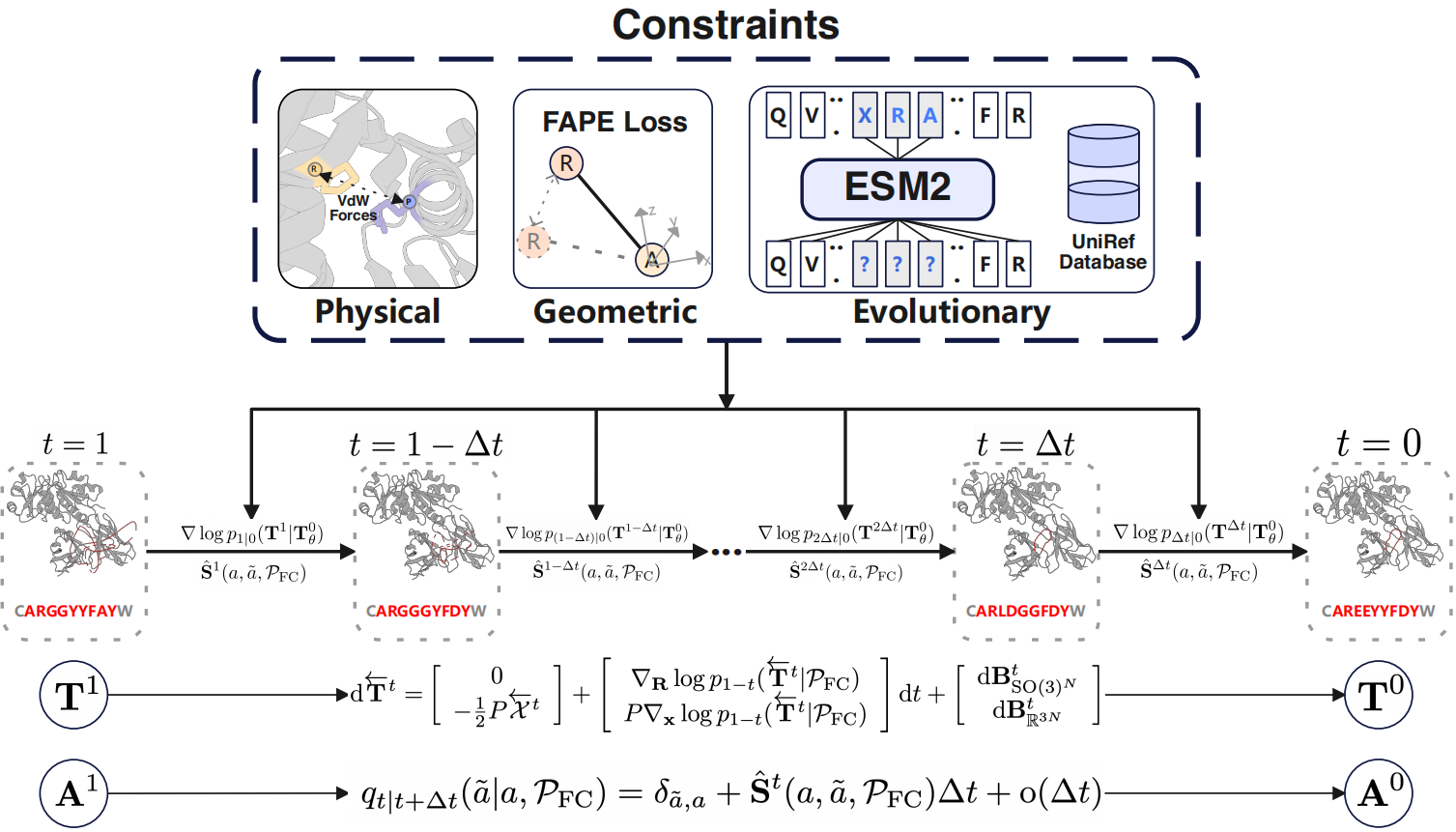
AbX: Antibody Design Using a Score-based Diffusion Model Guided by Evolutionary, Physical, and Geometric Constraints
Tian Zhu, Milong Ren, Haicang Zhang
Link to Paper at ICML 2024
If you encounter any issues with the installation or would like to report a bug, please feel free to open an issue on GitHub at https://github.com/CarbonMatrixLab/AbX/issues.
To install AbX, it is recommended to create a Conda environment and install the necessary dependencies by following these steps:
git clone [email protected]:CarbonMatrixLab/AbX.git
conda env create -f environment.yml
pip install fair-esm PyRosetta is required to relax the generated structures and compute binding energy. Please refer to the installation guide provided here for further instructions.
Antibody-antigen structures and associated summary files can be retrieved from the SAbDab database. The dataset and accompanying files can be downloaded from the following links:
Extract all_structures.zip into the data directory.
To preprocess the structure data into .npz format, use the preprocess_data.py script:
python preprocess_data.py --cpu 100 --summary_file ./data/sabdab_summary_all.tsv --data_dir ./data/mmcif --output_dir ./data/npz --data_mode mmcifWe recommend using the mmCIF format for PDB structures, as it provides comprehensive information.
- Download the AbX-DiffAb and AbX-RAbD model weights , and place them in the
./trained_modeldirectory. - Download the ESM2 model weights from here and the contact regressor weights from here, and save these files in the
./trained_modeldirectory.
To perform co-design of CDRs using the DiffAb test dataset, use the following command:
CUDA_VISIBLE_DEVICES=0 python inference.py \
--model ./trained_model/abx_diffab.ckpt \
--model_features ./config/config_data_feature.json \
--model_config ./config/config_model.json \
--batch_size 1 \
--num_samples 100 \
--name_idx ./test_data/diffab_test.idx \
--data_dir ./data/npz \
--output_dir ./output/DiffAb_design \
--mode designFor co-design using the RAbD test dataset, execute the following:
CUDA_VISIBLE_DEVICES=0 python inference.py \
--model ./trained_model/abx_rabd.ckpt \
--model_features ./config/config_data_feature.json \
--model_config ./config/config_model.json \
--batch_size 1 \
--num_samples 100 \
--name_idx ./test_data/RAbD_test.idx \
--data_dir ./data/npz \
--output_dir ./output/RAbD_design \
--mode designTo optimize CDRs in the DiffAb test dataset, run the following command:
CUDA_VISIBLE_DEVICES=0 python inference.py \
--model ./trained_model/abx_diffab.ckpt \
--model_features ./config/config_data_feature.json \
--model_config ./config/config_model.json \
--batch_size 1 \
--num_samples 100 \
--name_idx ./test_data/diffab_test.idx \
--data_dir ./data/npz \
--output_dir ./output/DiffAb_optimize \
--mode optimizeModify the generate_area and optimize_steps parameters to adjust the target regions and optimization steps.
To generate a trajectory during the design of CDRs in the DiffAb test dataset, use the following:
CUDA_VISIBLE_DEVICES=0 python inference.py \
--model ./trained_model/abx_diffab.ckpt \
--model_features ./config/config_data_feature.json \
--model_config ./config/config_model.json \
--batch_size 1 \
--num_samples 100 \
--name_idx ./test_data/diffab_test.idx \
--data_dir ./data/npz \
--output_dir ./output/DiffAb_optimize \
--mode trajectoryTo generate CDRs of given antibdody-antigen complexes in the PDB format, use the following:
CUDA_VISIBLE_DEVICES=0 python design.py \
--model ./trained_model/abx_diffab.ckpt \
--model_features ./config/config_data_feature.json \
--model_config ./config/config_model.json \
--batch_size 1 \
--num_samples 100 \
--pdb_file ./test_data/6ct7_H_L_S.pdb \
--output_dir ./output/design \
--mode designThe example of input antibody-antigen complexes is 6ct7_H_L_S.pdb, where H is the heavy chain id, L is the light chain id and S is the antigen chain id.
To relax the designed proteins using PyRosetta, run the following command and modify the relaxation regions using the generate_area parameter:
CUDA_VISIBLE_DEVICES=0 python relax_pdb.py \
--data_dir ./output/output_dir \
--cpus 100 \
--generate_area cdrsTo compute the RMSD, AAR, and IMP metrics, use the eval_metric.py script as follows:
CUDA_VISIBLE_DEVICES=0 python eval_metric.py \
--data_dir ./output/output_dir \
--cpus 100 \
--energyFor calculating plausibility, you may use AntiBERTy.
@inproceedings{
zhu2024antibody,
title={Antibody Design Using a Score-based Diffusion Model Guided by Evolutionary, Physical and Geometric Constraints},
author={Tian Zhu and Milong Ren and Haicang Zhang},
booktitle={Forty-first International Conference on Machine Learning},
year={2024},
url={https://openreview.net/forum?id=1YsQI04KaN}
}