iDrug is a computational pipeline to jointly predict novel drug-disease and drug-target interactions based on a heterogeneous network. iDrug adpots cross-network embedding to learn lower dimensional feature spaces for drugs, targets, and diseases in the heterogeneous network. A paper describing the approach is currently under review. A link to the paper will be added once it is published.
Drug-Disease Interactions: the drug-disease interactions in form of(DrugBankID, OMIMID)were downloaded from CTD database (https://ctdbase.org/downloads/;jsessionid=0CF3C56EC170EF21331BFCDFA5E230C0).Drug-Target Interactions: the drug-target interactions in form of(DrugBankID, UniprotID)were downloaded from DrugBank database (https://www.drugbank.ca/releases/latest).DiseaseName: disease names with theirOMIMIDin the drug-disease domain.DrugDomain1: drug names with theirDrugBankIDin the drug-disease domain.DrugDomain2: drug names with theirDrugBankIDin the drug-target domain.Target: target names with theirUniprotIDin the drug-target domain.
DrugDisease.mat: drug-disease interactions.DrugTarget.mat: drug-target interactions.DrugSimMat1.mat: drug-drug similarities in drug-disease domain.DrugSimMat2.mat: drug-drug similarities in drug-target domain.DiseaseSimMat.mat: disease-disease similarities.TargetSimMat.mat: target-target similarities.SMat.mat: the mapping matrix to denote the anchor links across the two domains.
- the histogram of similarity scores in the drug-disease domain.
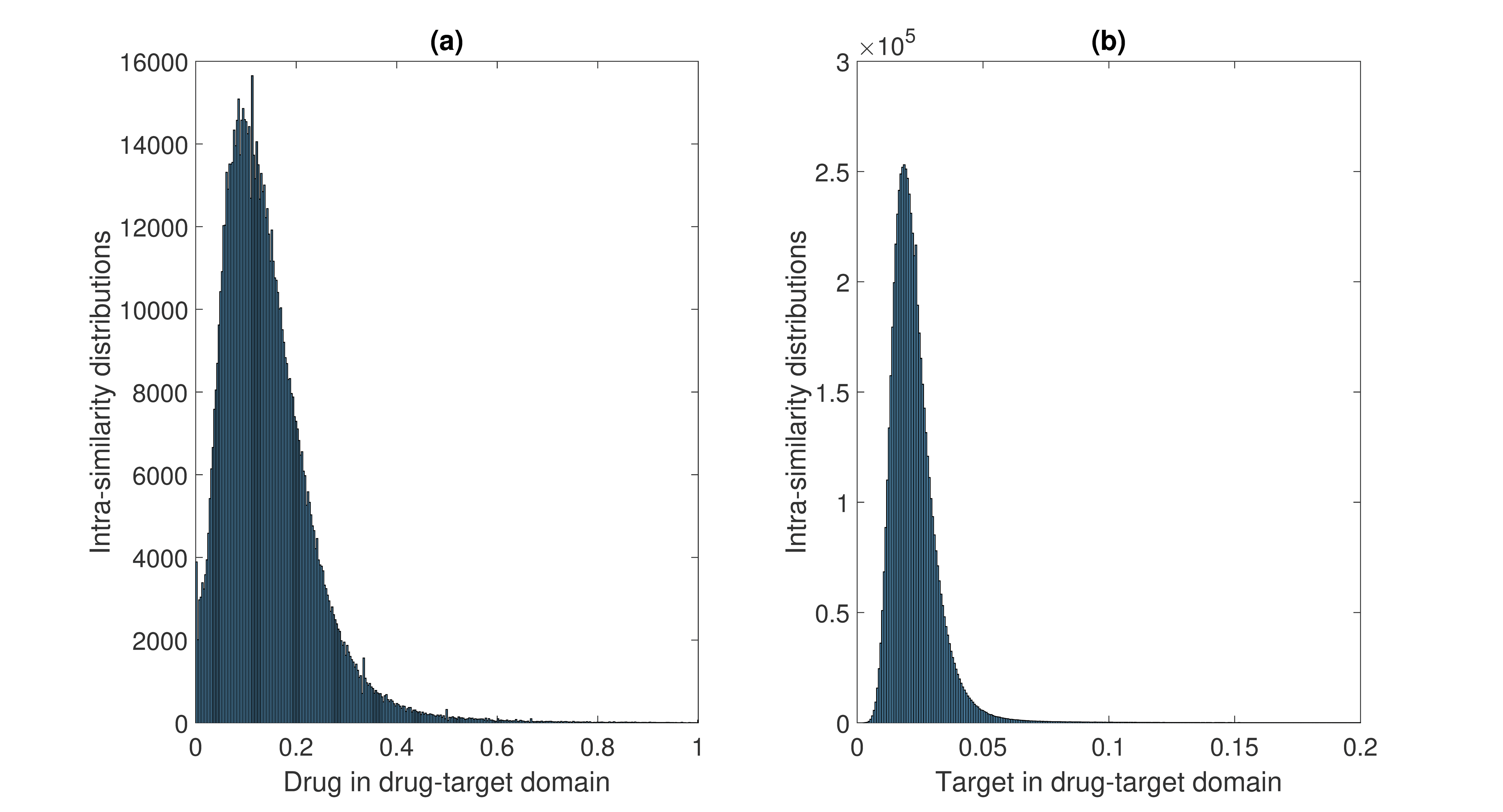
- the histogram of similarity scores in the drug-target domain.
iDrug.m: the optimization algorithm for iDrug framework.main.m: demo code for runningiDrug.m.train_test_split.m: split the data into training and testing sets.auc.m: evaluation script for AUROC and AUPR measurements.
- The code is tested under
MATLAB2015b.
We provide an example script to run experiments on our datasets:
- Users can run
main.mto replicate results in the paper.
matlab main(rank1, rank2, w, alpha, beta, gamma, DorT, scenario, k)
Among the parameters, rank1 and rank2 are the ranks of the latent matrices, w is the cost associated with the unobserved samples, and (alpha, beta, gamma) represent the contributions of within-domain smoothness, cross-network consistency, and the sparseness of solutions, respectively. The detail explanation of these parameters and sensitivity analysis can be found in the paper. The parameter DorT indicates whether the experiments will be for Drug-Disease (DorT = '1') or Drug-target (DorT = '2') prediction. The parameter scenario indicates the one of the three possible scenarios in the paper: pair prediction (scenario='1'), new drug (scenario='2'), or new disease (scenario='3'). For scenario 1, by varying the rank threshold, the algotirhn can calculate various true positive rate (TPR), false positive rate (FPR), Precision and Recall values. Area Under the Receiver Operating Characteristic curve (AUROC) and Area Under the Precision Recall curve (AUPR) will be generated. For senarios 2 and 3, the algorithm focuses on the performance of the top-k predictions, users need to set a value for the parameter k, and the algorithm output the precision calculated based on the top-k predictions. Parameter k is not used for scenario='1'.
To replicate drug-disease interaction prediction results using five-fold cross-validation in our paper, the values of these parameters are: rank1 = 90, rank2 = 70, w = 0.3, alpha=beta=gamma= 0.001, DorT = '1', scenario = '1'. The results using these parameters are shown here:
To replicate the new drug and new disease prediction in the paper, users can call the main function using the following parameters: matlab main(70, 70, 0.3, 0.01, 0.01, 0.01, 1, '2', 20). The output of the algorithm is the precision: precision: 0.23
The Gold Standard datasets are located in the ./goldDataset/ directory and the same variable names are used to store the interactions and similarities. To obtain results from the gold starndard data, users just need to change their working directory to ./goldDataset/.
Users just need to construct their data in Matlab format using the same name as outlined in the session "Data in MatLab form".
- RLS-Kron: http://cs.ru.nl/~tvanlaarhoven/drugtarget2011/
- MBiRW: https://github.com//bioinfomaticsCSU/MBiRW
- GRMF and TH_HGBI: we implemented these two algorithms in MatLab in the files
GRMF.mandTH_HGBI.mrespectively.
If you have any questions or comments, please feel free to email Huiyuan Chen (hxc501[at]case[dot]com).