BINN documentation is avaiable here.
The BINN-package allows you to create a sparse neural network from a pathway and input file. The examples presented in docs use the Reactome pathway database and a proteomic dataset to generate the neural network. It also allows you to train and interpret the network using SHAP. Plotting functions are also available for generating sankey plots. The article presenting the BINN can currently be found here.
Have a look at the poster_ndpia.ipynb for an example of a complete quick and easy BINN analysis.
BINN can be installed via pip
pip install binn
The package can also be built from source and installed with git.
git clone [email protected]:InfectionMedicineProteomics/BINN.git
pip install -e BINN/
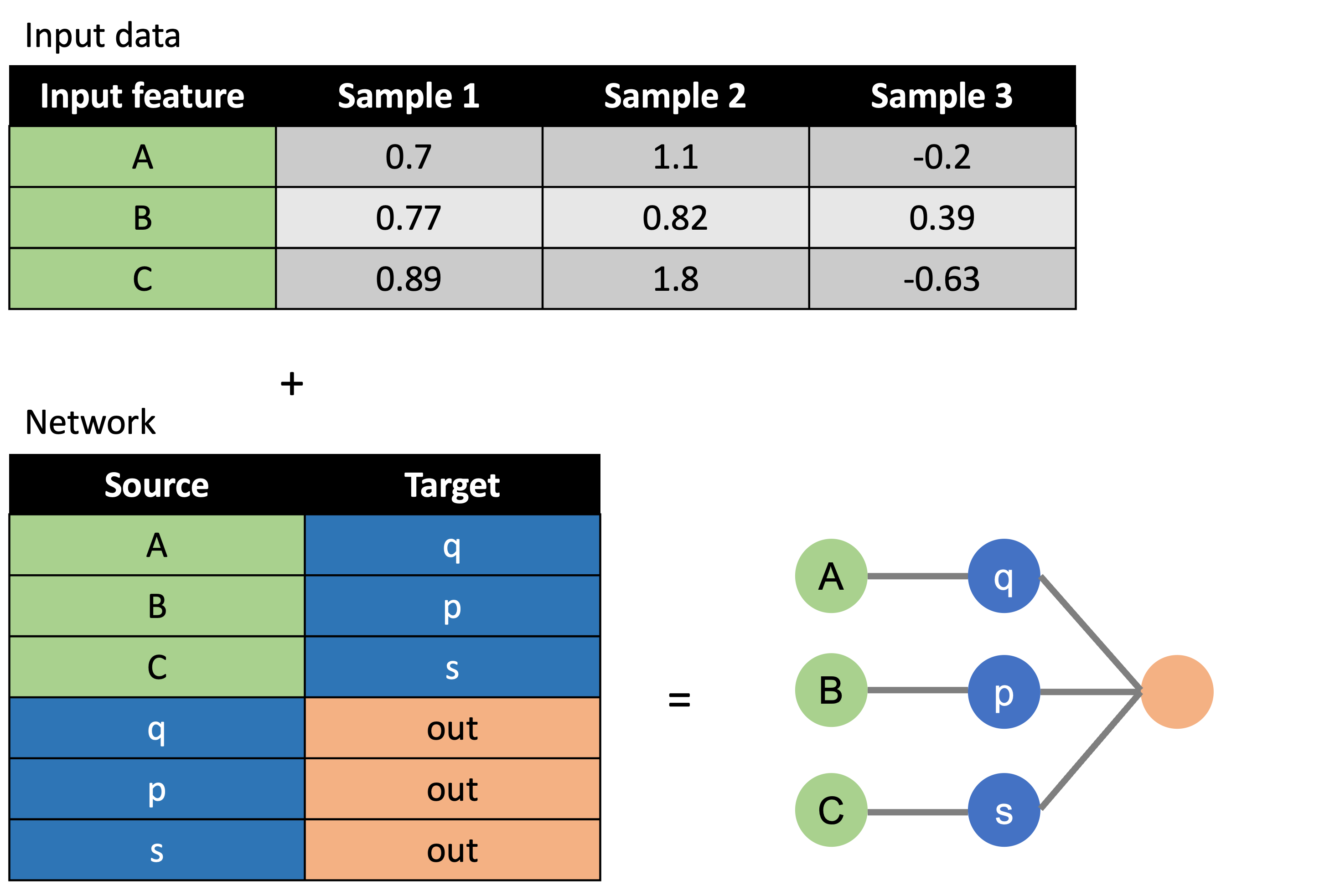
First, a network is created. This is the network that will be used to create the sparse BINN.
from binn import BINN, Network
import pandas as pd
input_data = pd.read_csv("../data/test_qm.tsv", sep="\t")
translation = pd.read_csv("../data/translation.tsv", sep="\t")
pathways = pd.read_csv("../data/pathways.tsv", sep="\t")
network = Network(
input_data=input_data,
pathways=pathways,
mapping=translation,
verbose=True
)The BINN can thereafter be generated using the network:
binn = BINN(
pathways=network,
n_layers=4,
dropout=0.2,
validate=False,
)An sklearn wrapper is also available:
from binn import BINNClassifier
binn = BINNClassifier(
pathways=network,
n_layers=4,
dropout=0.2,
validate=True,
epochs=10,
threads=10,
)This generates the Pytorch sequential model:
Sequential(
(Layer_0): Linear(in_features=446, out_features=953, bias=True)
(BatchNorm_0): BatchNorm1d(953, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(Dropout_0): Dropout(p=0.2, inplace=False)
(Tanh 0): Tanh()
(Layer_1): Linear(in_features=953, out_features=455, bias=True)
(BatchNorm_1): BatchNorm1d(455, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(Dropout_1): Dropout(p=0.2, inplace=False)
(Tanh 1): Tanh()
(Layer_2): Linear(in_features=455, out_features=162, bias=True)
(BatchNorm_2): BatchNorm1d(162, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(Dropout_2): Dropout(p=0.2, inplace=False)
(Tanh 2): Tanh()
(Layer_3): Linear(in_features=162, out_features=28, bias=True)
(BatchNorm_3): BatchNorm1d(28, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(Dropout_3): Dropout(p=0.2, inplace=False)
(Tanh 3): Tanh()
(Output layer): Linear(in_features=28, out_features=2, bias=True)
)Data - this file should contain a column with the feature names (quantmatrix or some matrix containing input column - in this case "Protein"). These need to map to the input layer of the BINN, either directly or by providing a translation file.
| Protein |
|---|
| P00746 |
| P00746 |
| P04004 |
| P27348 |
| P02751 |
| ... |
Pathways file - this file should contain the mapping used to create the connectivity in the hidden layers.
| target | source |
|---|---|
| R-BTA-109581 | R-BTA-109606 |
| R-BTA-109581 | R-BTA-169911 |
| R-BTA-109581 | R-BTA-5357769 |
| R-BTA-109581 | R-BTA-75153 |
| R-BTA-109582 | R-BTA-140877 |
| ... |
Translation file - this file is alternative, but is useful if some translation is needed to map the input features to the pathways in the hiddenn layers. In this case, it is used to map proteins (UniProt IDs) to pathways (Reactome IDs).
| input | translation |
|---|---|
| A0A075B6P5 | R-HSA-166663 |
| A0A075B6P5 | R-HSA-173623 |
| A0A075B6P5 | R-HSA-198933 |
| A0A075B6P5 | R-HSA-202733 |
| A0A075B6P5 | R-HSA-2029481 |
| ... |
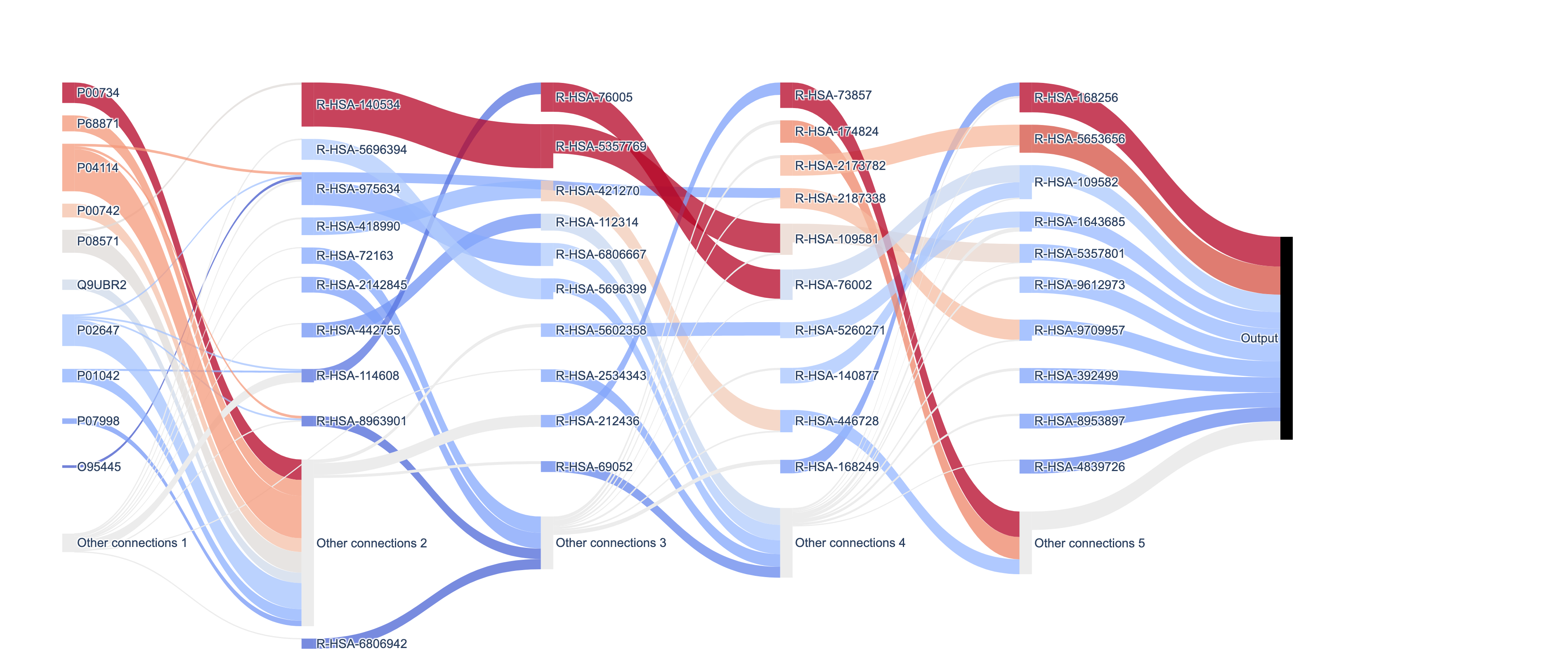
Plotting a subgraph starting from a node generates the plot:
 A complete sankey may look like this:
A complete sankey may look like this:
The software has been tested on desktop machines running Windows 10/Linux (Ubuntu). Small networks are not RAM-intensive and all experiments have been run comfortably with 16 GB RAM.
Please cite:
Hartman, E., Scott, A.M., Karlsson, C. et al. Interpreting biologically informed neural networks for enhanced proteomic biomarker discovery and pathway analysis. Nat Commun 14, 5359 (2023). https://doi.org/10.1038/s41467-023-41146-4
if you use this package.
Erik Hartman, infection medicine proteomics, Lund University
Aaron Scott, infection medicine proteomics, Lund University
Erik Hartman - [email protected]