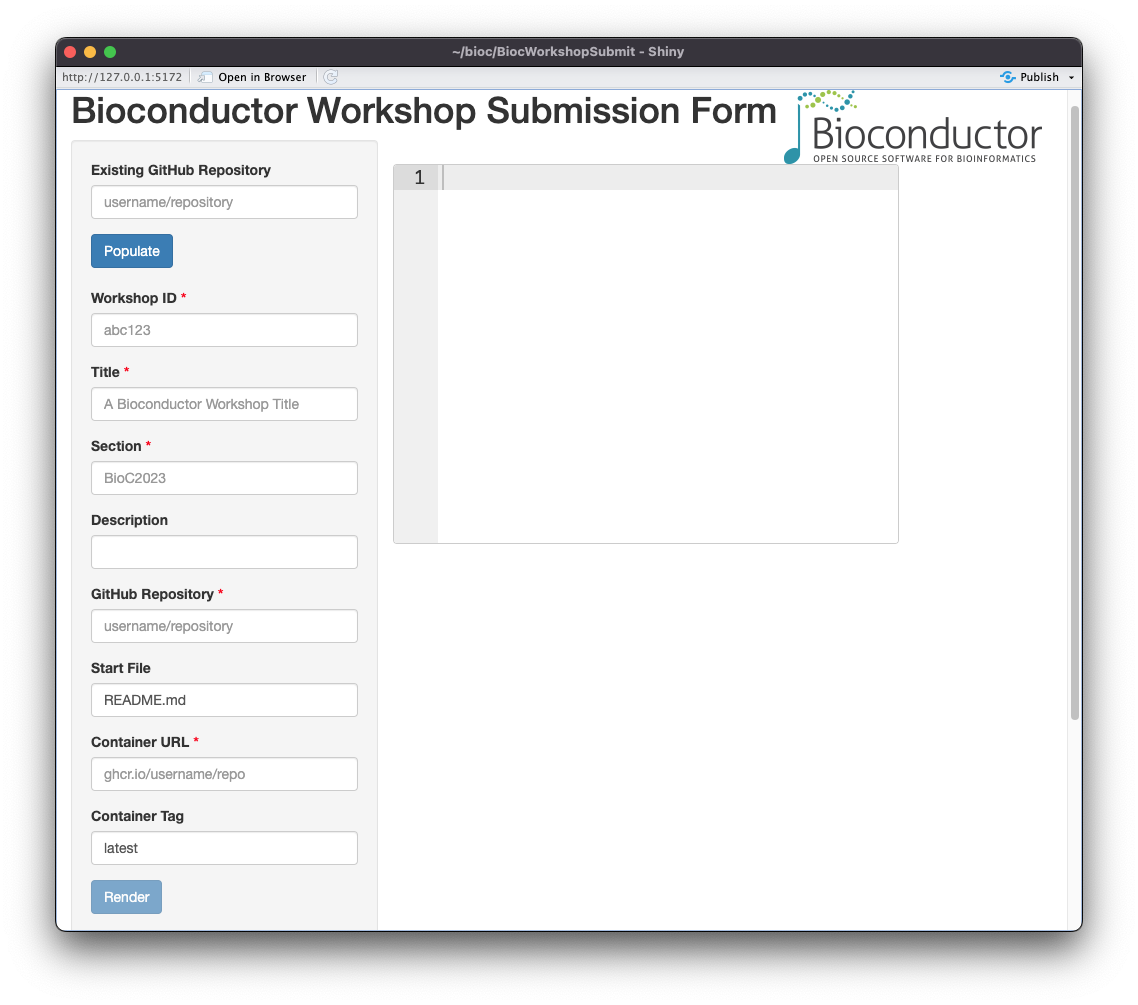
The BiocWorkshopSubmit package is a Shiny app that allows users to
submit Bioconductor workshops to the Bioconductor
Workshop website via the workshop
contributions repository on
GitHub. The app
is designed to be used as a standalone local app with a particular
setup.
The package is only available on GitHub and can be installed with the following command:
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("Bioconductor/BiocWorkshopSubmit")Workshop submitters should include the following files in their workshop:
A DESCRIPTION file with the following fields:
- Title: A title for the workshop
- Description: A short description of the workshop
- URL: A URL to the workshop repository from either GHCR or Docker Hub
Note. The container should be built using the Dockerfile that is
included in the example
BuildABiocWorkshop
repository. It can either point to docker.io or ghcr.io.
The DESCRIPTION file should be used to indicate what packages are
needed for the workshop. The DESCRIPTION file should also be used to
indicate what version of R is needed for the workshop.
Once the container is built, the workshop should be tested locally to ensure that it works as expected. The workshop should be tested (preferably) on the latest release version of R and Bioconductor.
The app will create a GitHub issue in the the workshop-contributions
repository. Submitters should generate a fine-grained personal access
token via GitHub https://github.com/settings/tokens. The token should
have the public_repo scope. The token should be saved with
gitcreds::gitcreds_set(). Once the token is saved, the app will use it
to create the issue.
The app can be launched with the following command:
BiocWorkshopSubmit::BiocWorkshopSubmit()The app will prompt the presenter to submit the following information:
- GitHub repository: The GitHub repository where the workshop contents are hosted.
- Workshop ID: A unique identifier for the workshop, e.g.,
tidybioc2023. - Section: The section that corresponds to the group of workshops for a
particular event (if any; e.g.,
Bioc2023). - Start File: The file that should be open when the user logs in to the workshop instance.
- Container Tag: The tag for the container that should be used for the
workshop (typically,
latest).
The workshop presenter will be able to auto-populate the fields in the
app by clicking the Populate button. The app will use the
DESCRIPTION file on GitHub to update the fields in the app.
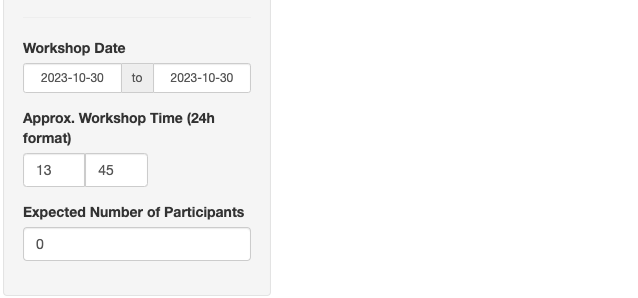
Workshop presenters can also provide additional information regarding the size, start time, and date of the workshop. This information will be used to allocate resources for the workshop on the Bioconductor Workshop website.
- Workshop Date: The date interval of the workshop event.
- Workshop Start Time (24h format): The start time of the workshop event in Eastern US time.
- Expected Number of Participants: The expected number of participants for the workshop.
After the details of the workshop have been entered, the workshop
presenter will be able to click the red Submit button to submit the
workshop to the workshop-contributions repository. The app will create
a GitHub issue in the workshop-contributions repository with the
details of the workshop. Please monitor the issue for any comments from
the Bioconductor team.
Thank you for your contribution to the Bioconductor Workshop website!
The following workshops have been submitted to the Bioconductor Workshop:
- MultiAssayWorkshop: https://github.com/waldronlab/MultiAssayWorkshop
- ISMB.OSCA: https://github.com/Bioconductor/ISMB.OSCA
Feel free to refer to these workshops as examples of how to structure the workshop package for submission to the Bioconductor Workshop website.