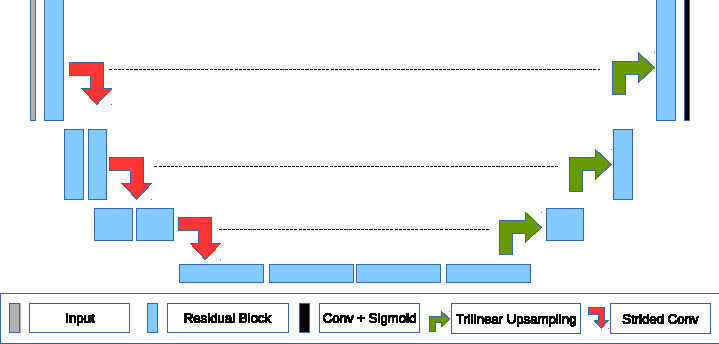
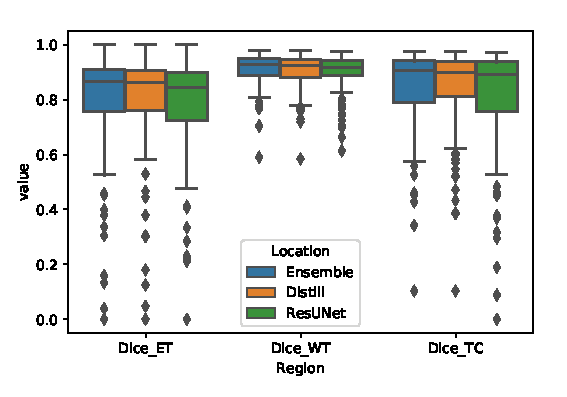
This code was written for participation in the Brain Tumor Segmentation Challenge (BraTS) 2019. The code is based on the corresponding paper, where we employ knowledge distillation for automatic brain tumor segmentation. First, we annotated unlabeled data with ensemble of different models. Then, we trained simple UNet model with residual blocks. This repository contains the code for inferencing the distilled model.
| Data | WT | ET | TC |
|---|---|---|---|
| Cascaded UNet Val 2019 | 0.9 | 0.731 | 0.833 |
| Distilled Val 2019 | 0.904 | 0.756 | 0.842 |
| Distilled Test 2019 | 0.887 | 0.794 | 0.829 |
Download checkpoint with the following link, where the model has the name brain-tumor-segmentation-0002.
You can also download onnx version for cpu inference with OpenVino.
The onnx model has a few modifications to match ONNX and OpenVino standards:
- It takes image of size
128x128x128as an input - It uses nearest neighbour upsampling instead of trilinear one
- It includes group norm operation from OpenVino
- Download BraTS data
- Download pretrained model
- Setup directory hierarchy
- Run the script
Run the following command. Models root directory is a repository for all models. Change the output directory in the script.
python test.py --name=brain-tumor-segmentation-0002 --models_path=<model's root dir>
dicom 0.9.9.post1
h5py 2.9.0
jupyter-client 5.2.4
jupyter-core 4.5.0
jupyterlab 1.0.1
jupyterlab-server 1.0.0
matplotlib 3.1.1
nibabel 2.4.1
numpy 1.16.4
opencv-python-headless 4.1.0.25
openpyxl 2.6.2
pandas 0.24.2
pickleshare 0.7.5
Pillow 6.1.0
protobuf 3.8.0
pydicom 1.2.2
Pygments 2.4.2
PyWavelets 1.0.3
scikit-image 0.15.0
scikit-learn 0.21.2
scipy 1.3.0
seaborn 0.9.0
SimpleITK 1.2.0
six 1.12.0
sklearn 0.0
tensorboardX 1.8
torch 1.2.0
torchvision 0.4.0
tqdm 4.32.2
TBD