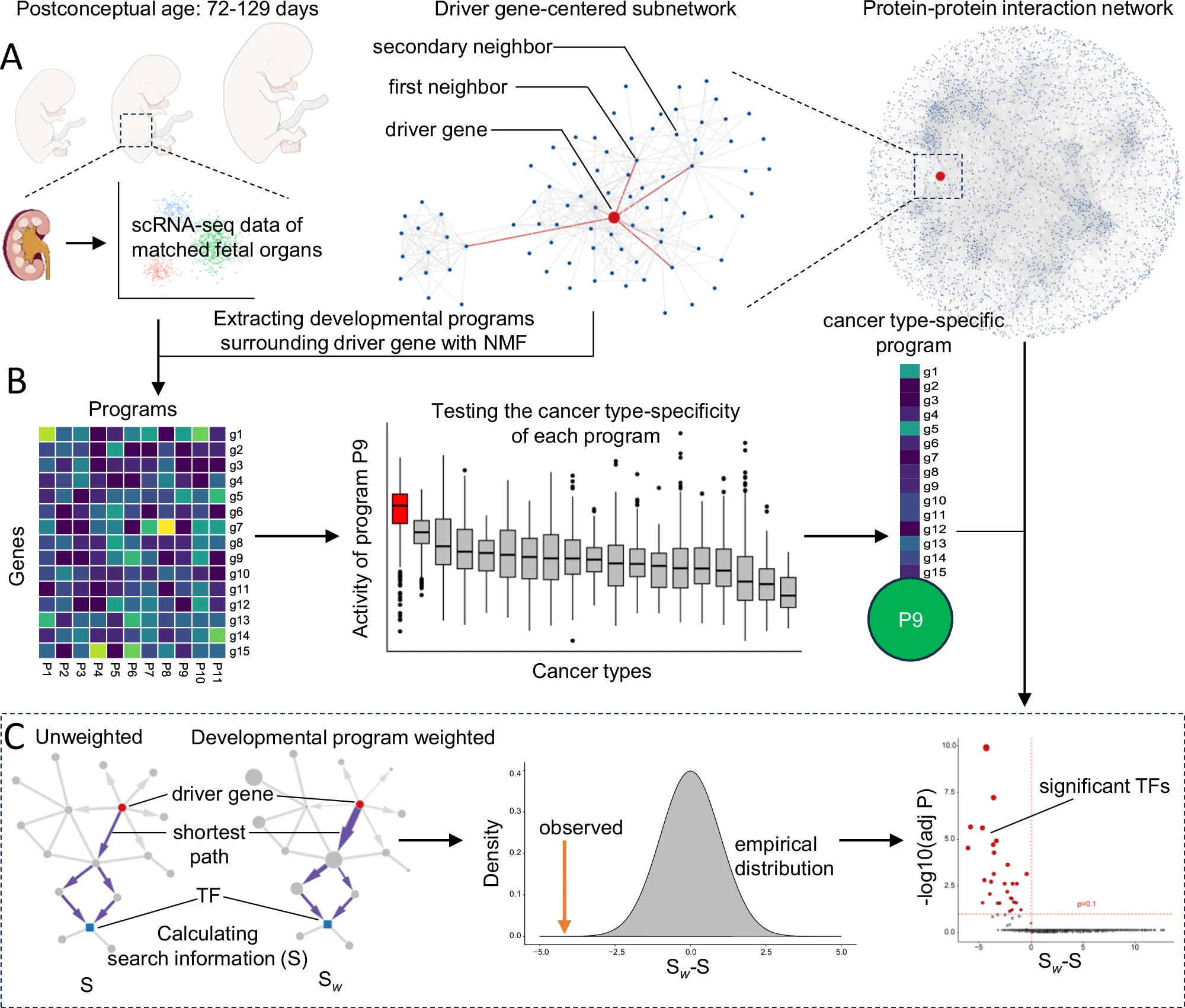
A computational framework for analyzing the link between development programs and cancer type-specific drivers via integrative network modeling. It combines single-cell gene expression profiles during fetal organ development, latent factor discovery, and information theory-based differential network analysis to systematically identify transcription factors that selectively respond to driver mutations under the influence of organ-specific developmental programs. This method could also be used to study other factors (such as wound healing, chronic inflammation or aging) that may influence the effects of driver mutations only if suitable scRNA-seq data are available. OncoNicheDev are written in Jupyter Notebook (Python) and R code, and details of how to use them can be found in the notes therein.
Identification of driver gene-centered protein-protein interaction subnetwork (code in subnetwork/)
Extracting developmental programs surrounding driver gene with NMF algorithm and testing the cancer type-specificity of each program (code in NMF/)
Comparing the effects of developmental programs on the efficiency of communication between driver genes and downstream transcription factors (code in network communication/)
Gene regulatory network analysis of influenced transcription factors (code in GRN/)
If you find our approach helpful, please cite:
Dong, X., et al. (2024). Network modeling links kidney developmental programs and the cancer type-specificity of VHL mutations. npj Systems Biology and Applications 10(1): 114.