Ramses A. Rosales-Garcia, Rhett M. Rautsaw , Erich P. Hofmann, Christoph I. Grunwald, Jason M. Jones, Hector Franz-Chavez, Ivan T. Ahumada-Carrillo, Ricardo Ramirez-Chaparro, Miguel Angel De la Torre-Loranca, Jason L. Strickland, Andrew J. Mason, Matthew L. Holding, Miguel Borja, Gamaliel Castaneda-Gaytan, Darin R. Rokyta, Tristan D. Schramer, N. Jade Mellor, Edward A. Myers, Christopher Parkinson 2023 April 03
Models were made following modeller basic tutorial using especifications from Whittington et al. (2018)
General pipeline was to print the translated sequence in ali format:
cat CgCrotB/CgCrotB.ali## >P1;CgCrotB
## sequence:CgCrotB:::::::0.00: 0.00
## MRTLWIVAVLLVVVEGNLLQFNKMIKLETKKNAVPFYSFYGCYCGWGGRGKPMDATDRCCFEHDCCYGKLTKCNTKSDIYSYSWKSGFIMCGKGSWCEEHICECDRIAAACLRRSLSTYKYGYMFYLDSYCKGPSEKC*
To set the right format you have to substitute the name of you protein and the sequence, but let the other stuff you have to set a * as stop at the end
The general pipeline is two python scripts taken from the modeller basic tutorial that are present in the bin file
ls bin## Evaluate_Model.py
## align2d.py
## build_profile.py
## erase_models.sh
## model-single.py
## plot_profile.py
## shebang_script.sh
I used parallel to make several models at once when the chanin of the reference was the same (CgCrotB, CgSCrotA, CgNCrotB).
the script should be run as follow
- Align2d.py -a < target_sequence.ali > -p < reference.pdf > -c < pdb chain >
- model-sigle.py -a <aligned_target2refence.ali > -k < reference + chain > -s < target name > model-single
parallel -a list -j 2 --verbose " echo {}
mkdir {}_1
cd {}_1
cp ../{}/{}.ali .
cp ../Crotoxin/3r0l.pdb .
python ../bin/align2d.py -a {}.ali -p 3r0l.pdb -c D
python ../bin/model-single.py -a {}-3r0lD.ali -k 3r0lD -s {} > model-single.log
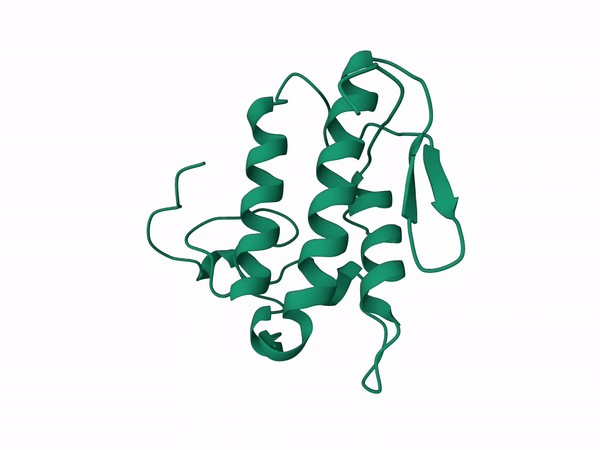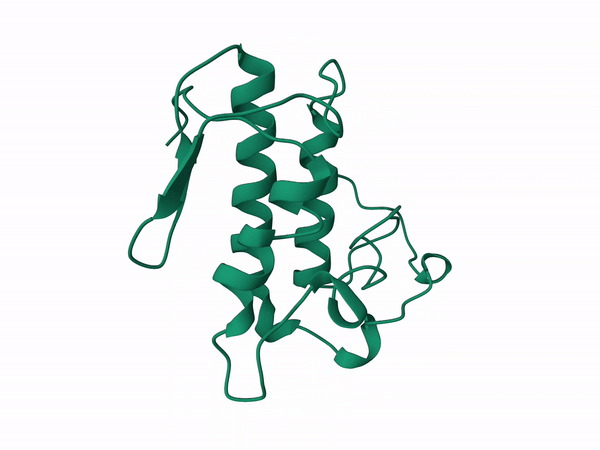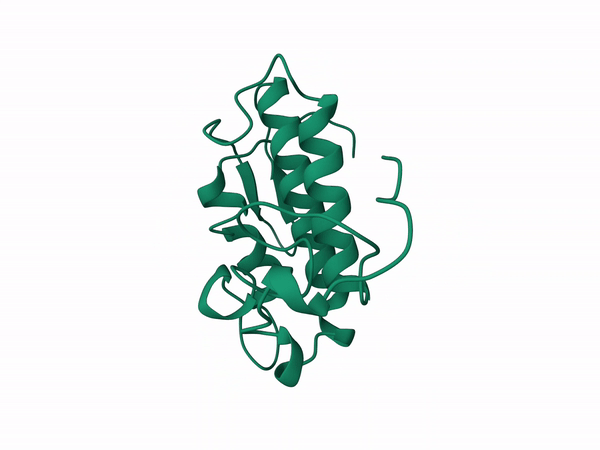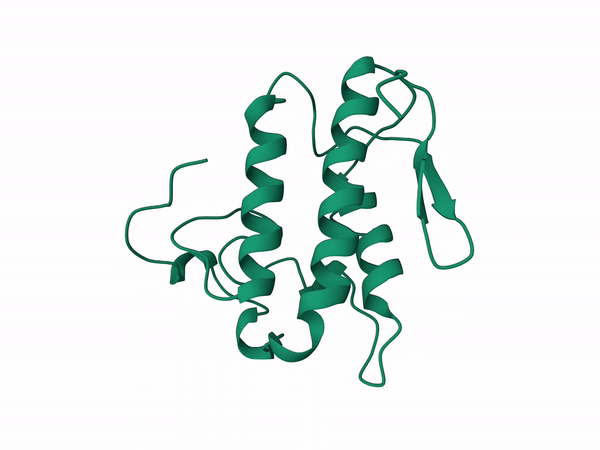
rm 3r0l.pdb"Beta subunit modeled with D chain of the crotoxin pdb corresponding to the B subunit.
Alfa subunit from the south population without cleavage modeled with D chain of the crotoxin pdb corresponding to the B subunit. The results from this run were used as input for GetArea server to calculate the Solvent Accesible Surface Area (SASA). The file used was CgSCrotA.B99990004.pdb, as it has the lowest DOPE-HR score, following Whittington et al. (2018)
tail -n 13 CgSCrotA/model-single.log## Filename molpdf DOPE-HR score GA341 score
## ----------------------------------------------------------------------
## CgSCrotA.B99990001.pdb 1046.83789 -6984.26514 0.99999
## CgSCrotA.B99990002.pdb 969.83252 -6598.30225 0.99999
## CgSCrotA.B99990003.pdb 1030.76172 -6981.58105 0.99999
## CgSCrotA.B99990004.pdb 970.64484 -7209.12451 1.00000
## CgSCrotA.B99990005.pdb 1256.93872 -5951.22266 0.99984
## CgSCrotA.B99990006.pdb 1048.81567 -6757.01074 0.99996
## CgSCrotA.B99990007.pdb 1003.13141 -6838.75195 0.99998
## CgSCrotA.B99990008.pdb 963.31897 -6584.64404 0.99996
## CgSCrotA.B99990009.pdb 957.17169 -6810.50195 1.00000
## CgSCrotA.B99990010.pdb 984.32172 -7163.36230 0.99998
Alfa subunit from the South population with cleavage on sites 1,4,5 modeled with D chain of the crotoxin pdb corresponding to the B subunit.
Alfa subunit from the South population with cleavage on sites 1,4,5 and 2,3 modeled with A,B,D chains of the crotoxin pdb corresponding to the A subunit.
Alfa subunit from the North population without cleavage modeled with D chain of the crotoxin pdb corresponding to the B subunit.
Alfa subunit from the North population with cleavage on sites 1,4,5 modeled with D chain of the crotoxin pdb corresponding to the B subunit.
Alfa subunit from the South population with cleavage on sites 1,4,5, and 2,3 modeled with A,B,D chains of the crotoxin pdb corresponding to the A subunit.