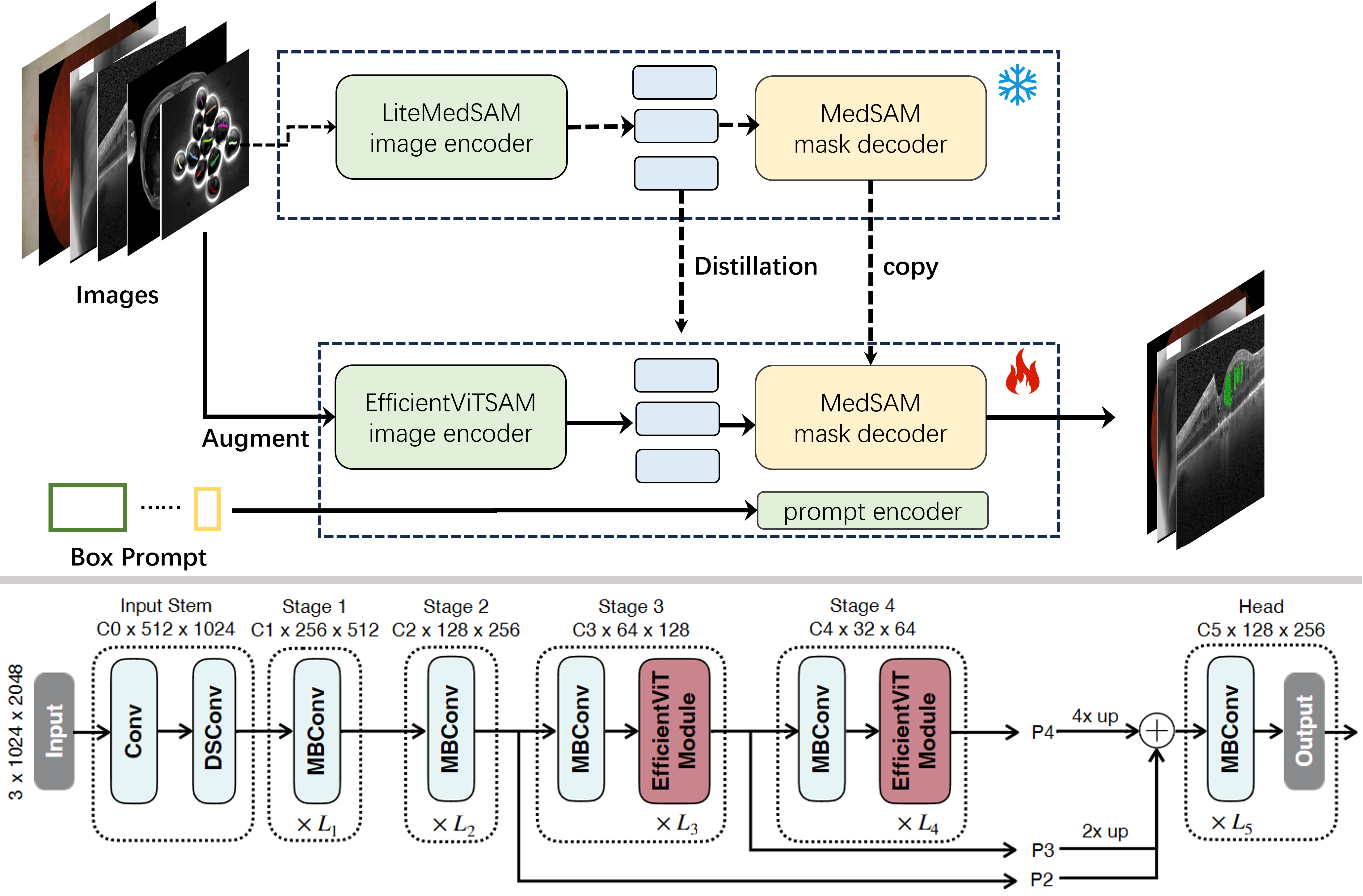
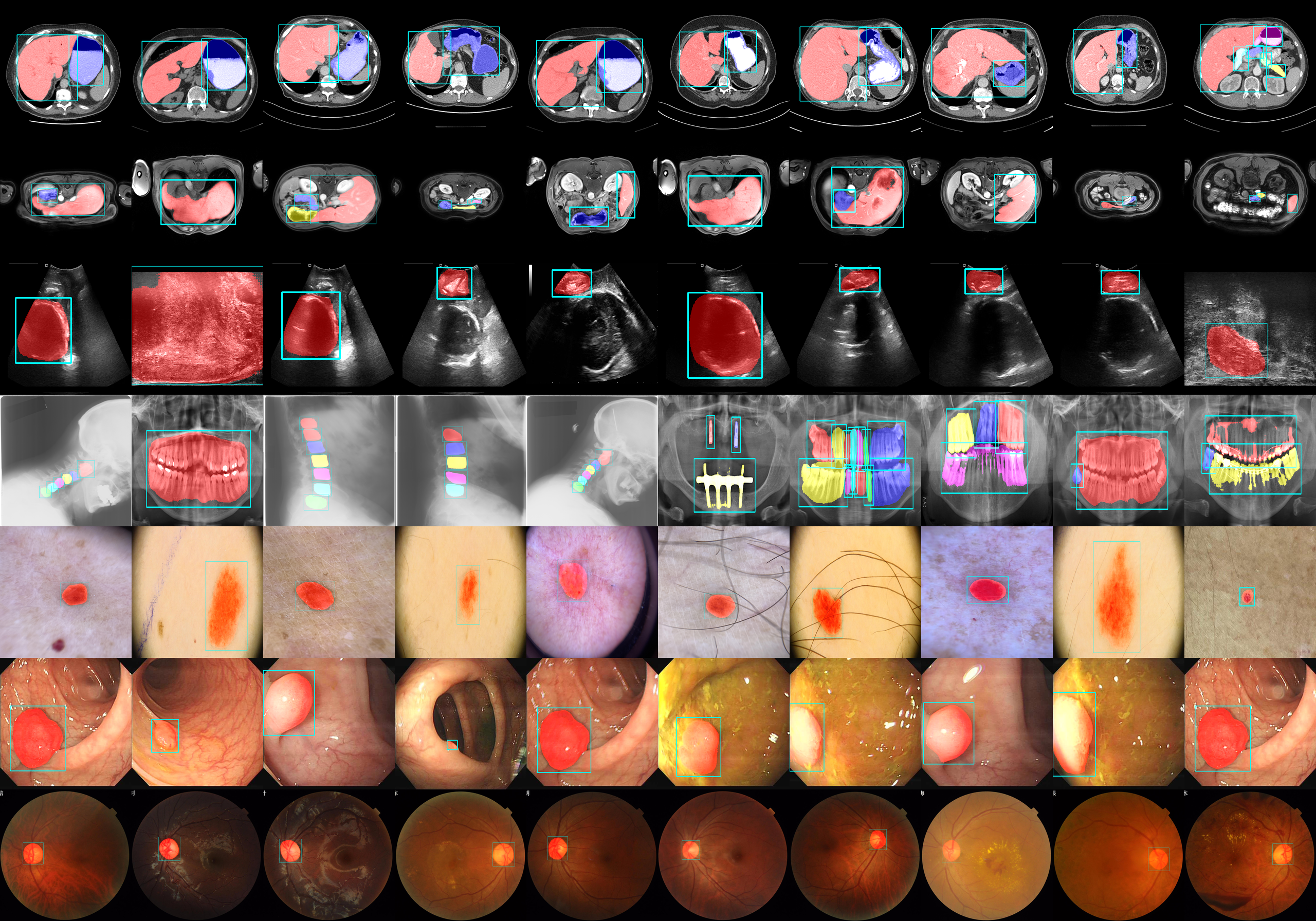
This repository is the implementation of reproduced medficientsam (Rank.1 Solution of CVPR Challenge-MedSAM Laptop).
| System | Ubuntu 22.04.6 LTS |
|---|---|
| CPU | Intel(R) Xeon(R) Silver 4114 |
| RAM | 128GB |
| GPU (number and type) | One NVIDIA 4080 16G |
| CUDA version | 12.1 |
| Python version | 3.10 |
| Deep learning framework | torch 2.2.2, torchvision 0.17.2 |
# 1. Clone repository and enter the folder
git clone https://github.com/RicoLeehdu/medficientsam-reproduce.git
cd medficientsam-reproduce
# 2. Create a virtual environment
conda env create -f environment.yaml -n medficientsam-reproduce
# 3. Activate it
conda activate medficientsam-reproduce
# 4. Install requirements by pip
pip install -r requirements.txt
# Optinal choice for accelerate pip installation
pip config set global.index-url https://mirrors.tuna.tsinghua.edu.cn/pypi/web/simpleThe Docker images can be found here.
# 1. Load image
docker load -i icimhdu_reproduce1st.tar.gz
# 2. Run test
docker container run -m 8G --name seno --rm -v $PWD/test_input/:/workspace/inputs/ -v $PWD/test_output/:/workspace/outputs/ icimhdu_reproduce1st:latest /bin/bash -c "sh predict.sh" To measure the running time (including Docker starting time), see https://github.com/bowang-lab/MedSAM/blob/LiteMedSAM/CVPR24_time_eval.py
-
Participate in the challenge to access the dataset the training and validation dataset.
-
Download and unzip the files to your dataset folder, copy
.env.exampleto.envand modifyCVPR2024_MEDSAM_DATA_DIRto the dataset path. The directory structure should look like this:CVPR24-MedSAMLaptopData ├── train_npz │ ├── CT │ ├── Dermoscopy │ ├── Endoscopy │ ├── Fundus │ ├── Mammography │ ├── Microscopy │ ├── MR │ ├── OCT │ ├── PET │ ├── US │ └── XRay ├── validation-box └── imgs
Note: There is no additional processing steps for training and validation dataset.
Model training is divided into two stages: distillation and fine-tuning, and model types are divided into three types: l0, l1 and l2.
-
Prepare pretrained weights from MedSAM .
Download the
medsam_vit_b.pthinto/weights/medsam. -
Start distill and fine-tune model.
# Take the l0 model type as an example: cd train_scripts # 1. Distillation l0 sh distill_l0.sh # 2. finetune l0 with augment sh finetune_l0_augment.sh # or finetune l0 without augment sh finetune_l0_unaugment.sh
Note: The data augment is to offset the position of the prompt box by a small amount to simulate the disturbance phenomenon of the prompt box.
More training configuration like batch_size and num_workers are defined in /configs/data/distill_medsam.yaml and /configs/data/finetune_medsam.yaml
# /configs/data/finetune_medsam.yaml
_target_: src.data.medsam_datamodule.MedSAMDataModule
train_val_test_split: [0.9, 0.05, 0.05]
batch_size: 32
num_workers: 32
pin_memory: true
dataset:
_target_: src.data.components.medsam_dataset.MedSAMTrainDataset
_partial_: true
data_dir: ${paths.cvpr2024_medsam_data_dir}/train_npz
image_encoder_input_size: 512
bbox_random_shift: 5
mask_num: 5
data_aug: true
scale_image: true
normalize_image: false
limit_npz: null
limit_sample: null
aug_transform:
_target_: albumentations.Compose
transforms:
- _target_: albumentations.HorizontalFlip
p: 0.5
- _target_: albumentations.VerticalFlip
p: 0.5
- _target_: albumentations.ShiftScaleRotate
shift_limit: 0.0625
scale_limit: 0.2
rotate_limit: 90
border_mode: 0
p: 0.5Modify checkpoint_path in yaml to the latest trained ckpt path.
# /config/experiment/finetune_l0.yaml
model:
model:
_target_: src.models.base_sam.BaseSAM.construct_from
original_sam:
_target_: src.models.segment_anything.build_sam_vit_b
checkpoint: ${paths.weights_dir}/medsam/medsam_vit_b.pth
distill_lit_module:
_target_: src.models.distill_module.DistillLitModule.load_from_checkpoint
checkpoint_path: ${paths.weights_dir}/distilled-l0/step_400000.ckpt
student_net:
_target_: src.models.efficientvit.sam_model_zoo.create_sam_model
name: l0
pretrained: false
teacher_net:
_target_: src.models.segment_anything.build_sam_vit_b
checkpoint: ${paths.weights_dir}/medsam/medsam_vit_b.pth-
Tool: Wandb
-
Method: configure the
WANDB_API_KEYin.env
-
Download trained checkpoints here and put them into individual folders, eg:
/medficientsam-reproduce/weights/distilled-l0. -
Modify
checkpoint_pathin yaml to the target ckpt path.# /config/experiment/finetune_l0.yaml model: _target_: src.models.base_sam.BaseSAM.construct_from original_sam: _target_: src.models.segment_anything.build_sam_vit_b checkpoint: ${paths.weights_dir}/medsam/medsam_vit_b.pth distill_lit_module: _target_: src.models.distill_module.DistillLitModule.load_from_checkpoint checkpoint_path: ${paths.weights_dir}/distilled-l0/step_400000.ckpt student_net: _target_: src.models.efficientvit.sam_model_zoo.create_sam_model name: l0 pretrained: false teacher_net: _target_: src.models.segment_anything.build_sam_vit_b checkpoint: ${paths.weights_dir}/medsam/medsam_vit_b.pth
-
Specify the experiment type and run the follow command. Take the finetuned l model type as an example:
python src/infer.py experiment=infer_finetuned_l1 \ ## inference type
Accuracy metrics are evaluated on the public validation set of CVPR 2024 Segment Anything In Medical Images On Laptop Challenge. The computational metrics are obtained on an Intel(R) Core(TM) i9-10900K.
| Method | Res. | Params | FLOPs | DSC | NSD | DSC-R | NSD-R | 2D Runtime | 3D Runtime | 2D Memory Usage | 3D Memory Usage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MedSAM | 1024 | 93.74M | 488.24G | 84.91 | 86.46 | 84.91 | 86.46 | N/A | N/A | N/A | N/A |
| LiteMedSAM | 256 | 9.79M | 39.98G | 83.23 | 82.71 | 83.23 | 82.71 | 5.1s | 42.6s | 1135MB | 1241MB |
| MedficientSAM-L0 | 512 | 34.79M | 36.80G | 85.85 | 87.05 | 84.93 | 86.76 | 0.9s | 7.4s | 448MB | 687MB |
| MedficientSAM-L1 | 512 | 47.65M | 51.05G | 86.42 | 87.95 | 85.16 | 86.68 | 1.0s | 9.0s | 553MB | 793MB |
| MedficientSAM-L2 | 512 | 61.33M | 70.71G | 86.08 | 87.53 | 85.07 | 86.63 | 1.1s | 11.1s | 663MB | 903MB |
| Target | LiteMedSAM DSC(%) | LiteMedSAM NSD(%) | Distillation DSC(%) | Distillation NSD(%) | Distillation-R DSC(%) | Distillation-R NSD(%) | No Augmentation DSC(%) | No Augmentation NSD(%) | No Augmentation-R DSC(%) | No Augmentation-R NSD(%) | MedficientSAM-L1 DSC(%) | MedficientSAM-L1 NSD(%) | MedficientSAM-L1-R DSC(%) | MedficientSAM-L1-R NSD(%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT | 92.26 | 94.90b | 91.13 | 93.75 | 92.15 | 94.74 | 92.24 | 94.71 | 92.69 | 95.50b | 92.15 | 94.80 | 93.19r | 95.78r |
| MR | 89.63r | 93.37r | 85.73 | 89.75 | 87.87b | 91.40 | 87.25 | 90.88 | 88.54 | 92.21b | 86.98 | 90.77 | 89.51 | 92.99 |
| PET | 51.58 | 25.17 | 70.49b | 54.52b | 68.30 | 50.17 | 72.05r | 56.26r | 61.06 | 49.13 | 73.00r | 58.03r | 66.97 | 52.52 |
| US | 94.77r | 96.81r | 84.43 | 89.29 | 84.52b | 89.37 | 81.99 | 86.74 | 82.41 | 87.16b | 82.50 | 87.24 | 81.39 | 86.09 |
| X-Ray | 75.83 | 80.39 | 78.92 | 84.64 | 75.40 | 80.38 | 79.88 | 85.73r | 78.04 | 83.10 | 80.47b | 86.23r | 75.78 | 80.88 |
| Dermoscopy | 92.47 | 93.85 | 92.84 | 94.16 | 92.54 | 93.88 | 94.24r | 95.62r | 93.71b | 95.19b | 94.16 | 95.54 | 93.17 | 94.62 |
| Endoscopy | 96.04b | 98.11 | 96.88r | 98.81r | 95.92 | 98.16 | 96.05 | 98.33 | 95.58 | 98.07b | 96.10 | 98.37 | 94.62 | 97.26 |
| Fundus | 94.81 | 96.41 | 94.10 | 95.83 | 93.85 | 95.54 | 94.16 | 95.89 | 94.27r | 96.00r | 94.32 | 96.05 | 94.16 | 95.90 |
| Microscopy | 61.63 | 65.38 | 75.63 | 82.15 | 75.90b | 82.45b | 78.76r | 85.22r | 78.09 | 84.48 | 78.09 | 84.47 | 77.67 | 84.11 |
| Average | 83.23 | 82.71 | 85.57 | 86.99 | 85.16 | 86.23 | 86.29r | 87.71 | 84.93 | 86.76 | 86.42 | 87.95r | 85.16 | 86.68 |
b - Suboptimal results marked in blue
- Distilled-L0
- Distilled-L1
- Distilled-L2
- Finetuned-L0
- Finetuned-L1
- Finetuned-L2
We thank the authors of MedSAM and medficientsam for making their source code publicly available and the organizer of CVPR Challenge-MedSAM on Laptop for their excellent work.