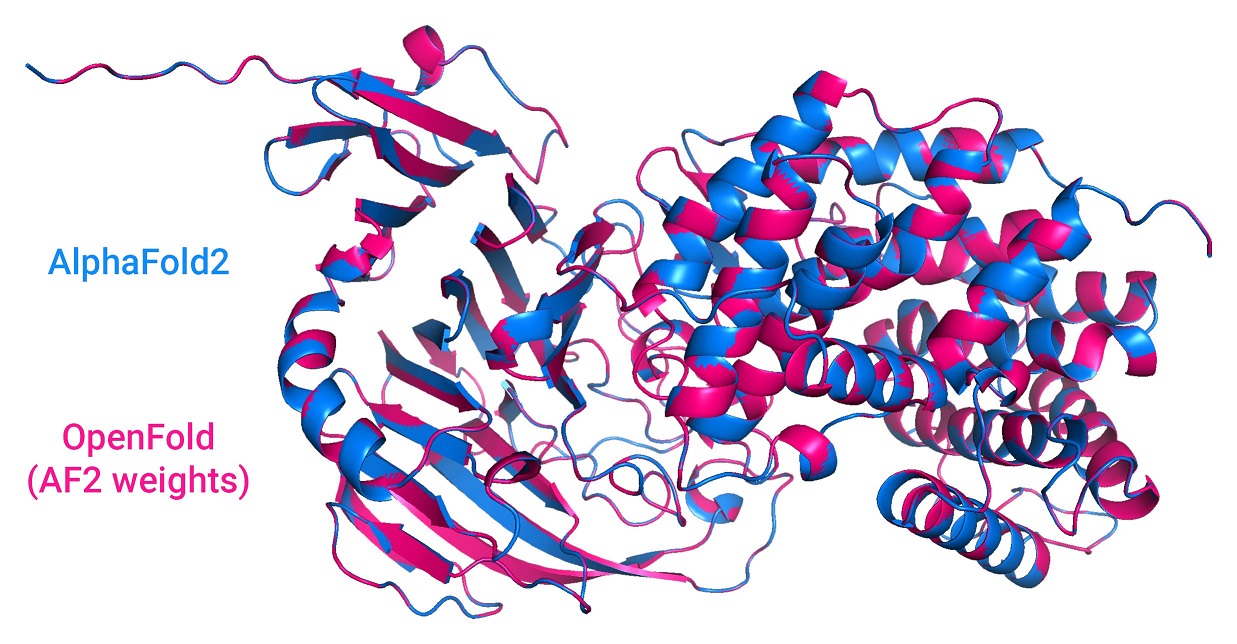
A faithful PyTorch reproduction of DeepMind's AlphaFold 2.
OpenFold carefully reproduces (almost) all of the features of the original open source inference code (v2.0.1). The sole exception is model ensembling, which fared poorly in DeepMind's own ablation testing and is being phased out in future DeepMind experiments. It is omitted here for the sake of reducing clutter. In cases where the Nature paper differs from the source, we always defer to the latter.
OpenFold is built to support inference with AlphaFold's original JAX weights. Try it out with our Colab notebook.
Unlike DeepMind's public code, OpenFold is also trainable. It can be trained
with DeepSpeed and with either fp16
or bfloat16 half-precision.
All Python dependencies are specified in environment.yml. For producing sequence
alignments, you'll also need kalign, the HH-suite,
and one of {jackhmmer, MMseqs2 (nightly build)}
installed on on your system. Finally, some download scripts require aria2c.
For convenience, we provide a script that installs Miniconda locally, creates a
conda virtual environment, installs all Python dependencies, and downloads
useful resources (including DeepMind's pretrained parameters). Run:
scripts/install_third_party_dependencies.shTo activate the environment, run:
source scripts/activate_conda_env.shTo deactivate it, run:
source scripts/deactivate_conda_env.shTo install the HH-suite to /usr/bin, run
# scripts/install_hh_suite.shTo download DeepMind's pretrained parameters and common ground truth data, run:
bash scripts/download_data.sh data/You have two choices for downloading protein databases, depending on whether you want to use DeepMind's MSA generation pipeline (w/ HMMR & HHblits) or ColabFold's, which uses the faster MMseqs2 instead. For the former, run:
bash scripts/download_alphafold_dbs.sh data/For the latter, run:
bash scripts/download_mmseqs_dbs.sh data/ # downloads .tar files
bash scripts/prep_mmseqs_dbs.sh data/ # unpacks and preps the databasesMake sure to run the latter command on the machine that will be used for MSA generation (the script estimates how the precomputed database index used by MMseqs2 should be split according to the memory available on the system).
Alternatively, you can use raw MSAs from
ProteinNet. After downloading
the database, use scripts/prep_proteinnet_msas.py to convert the data into
a format recognized by the OpenFold parser. The resulting directory becomes the
alignment_dir used in subsequent steps. Use scripts/unpack_proteinnet.py to
extract .core files from ProteinNet text files.
For both inference and training, the model's hyperparameters can be tuned from
openfold/config.py. Of course, if you plan to perform inference using
DeepMind's pretrained parameters, you will only be able to make changes that
do not affect the shapes of model parameters. For an example of initializing
the model, consult run_pretrained_openfold.py.
To run inference on a sequence or multiple sequences using a set of DeepMind's pretrained parameters, run e.g.:
python3 run_pretrained_openfold.py \
target.fasta \
data/pdb_mmcif/mmcif_files/ \
--uniref90_database_path data/uniref90/uniref90.fasta \
--mgnify_database_path data/mgnify/mgy_clusters_2018_12.fa \
--pdb70_database_path data/pdb70/pdb70 \
--uniclust30_database_path data/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
--output_dir ./ \
--bfd_database_path data/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--model_device cuda:1 \
--jackhmmer_binary_path lib/conda/envs/openfold_venv/bin/jackhmmer \
--hhblits_binary_path lib/conda/envs/openfold_venv/bin/hhblits \
--hhsearch_binary_path lib/conda/envs/openfold_venv/bin/hhsearch \
--kalign_binary_path lib/conda/envs/openfold_venv/bin/kalignwhere data is the same directory as in the previous step. If jackhmmer,
hhblits, hhsearch and kalign are available at the default path of
/usr/bin, their binary_path command-line arguments can be dropped.
If you've already computed alignments for the query, you have the option to
skip the expensive alignment computation here.
Note that chunking (as defined in section 1.11.8 of the AlphaFold 2 supplement)
is enabled by default in inference mode. To disable it, set globals.chunk_size
to None in the config.
After activating the OpenFold environment with
source scripts/activate_conda_env.sh, install OpenFold by running
python setup.py installTo train the model, you will first need to precompute protein alignments.
You have two options. You can use the same procedure DeepMind used by running the following:
python3 scripts/precompute_alignments.py mmcif_dir/ alignment_dir/ \
data/uniref90/uniref90.fasta \
data/mgnify/mgy_clusters_2018_12.fa \
data/pdb70/pdb70 \
data/pdb_mmcif/mmcif_files/ \
data/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
--bfd_database_path data/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--cpus 16 \
--jackhmmer_binary_path lib/conda/envs/openfold_venv/bin/jackhmmer \
--hhblits_binary_path lib/conda/envs/openfold_venv/bin/hhblits \
--hhsearch_binary_path lib/conda/envs/openfold_venv/bin/hhsearch \
--kalign_binary_path lib/conda/envs/openfold_venv/bin/kalignAs noted before, you can skip the binary_path arguments if these binaries are
at /usr/bin. Expect this step to take a very long time, even for small
numbers of proteins.
Alternatively, you can generate MSAs with the ColabFold pipeline (and templates with HHsearch) with:
python3 scripts/precompute_alignments_mmseqs.py input.fasta \
data/mmseqs_dbs \
uniref30_2103_db \
alignment_dir \
~/MMseqs2/build/bin/mmseqs \
/usr/bin/hhsearch \
--env_db colabfold_envdb_202108_db
--pdb70 data/pdb70/pdb70where input.fasta is a FASTA file containing one or more query sequences. To
generate an input FASTA from a directory of mmCIF and/or ProteinNet .core
files, we provide scripts/data_dir_to_fasta.py.
Next, generate a cache of certain datapoints in the template mmCIF files:
python3 scripts/generate_mmcif_cache.py \
mmcif_dir/ \
mmcif_cache.json \
--no_workers 16This cache is used to pre-filter templates.
Next, generate a separate chain-level cache with data used for training-time data filtering:
python3 scripts/generate_chain_data_cache.py \
mmcif_dir/ \
chain_data_cache.json \
--cluster_file clusters-by-entity-40.txt \
--no_workers 16where the cluster_file argument is a file of chain clusters, one cluster
per line (e.g. PDB40).
Finally, call the training script:
python3 train_openfold.py mmcif_dir/ alignment_dir/ template_mmcif_dir/ \
2021-10-10 \
--template_release_dates_cache_path mmcif_cache.json \
--precision 16 \
--gpus 8 --replace_sampler_ddp=True \
--seed 42 \ # in multi-gpu settings, the seed must be specified
--deepspeed_config_path deepspeed_config.json \
--checkpoint_every_epoch \
--resume_from_ckpt ckpt_dir/ \
--train_chain_data_cache_path chain_data_cache.jsonwhere --template_release_dates_cache_path is a path to the mmCIF cache.
A suitable DeepSpeed configuration file can be generated with
scripts/build_deepspeed_config.py. The training script is
written with PyTorch Lightning
and supports the full range of training options that entails, including
multi-node distributed training. For more information, consult PyTorch
Lightning documentation and the --help flag of the training script.
Note that the data directory can also contain PDB files previously output by the model. These are treated as members of the self-distillation set and are subjected to distillation-set-only preprocessing steps.
To run unit tests, use
scripts/run_unit_tests.shThe script is a thin wrapper around Python's unittest suite, and recognizes
unittest arguments. E.g., to run a specific test verbosely:
scripts/run_unit_tests.sh -v tests.test_modelCertain tests require that AlphaFold (v2.0.1) be installed in the same Python
environment. These run components of AlphaFold and OpenFold side by side and
ensure that output activations are adequately similar. For most modules, we
target a maximum pointwise difference of 1e-4.
Openfold can be built as a docker container using the included dockerfile. To build it, run the following command from the root of this repository:
docker build -t openfold .The built container contains both run_pretrained_openfold.py and train_openfold.py as well as all necessary software dependencies. It does not contain the model parameters, sequence, or structural databases. These should be downloaded to the host machine following the instructions in the Usage section above.
The docker container installs all conda components to the base conda environment in /opt/conda, and installs openfold itself in /opt/openfold,
Before running the docker container, you can verify that your docker installation is able to properly communicate with your GPU by running the following command:
docker run --rm --gpus all nvidia/cuda:11.0-base nvidia-smiNote the --gpus all option passed to docker run. This option is necessary in order for the container to use the GPUs on the host machine.
To run the inference code under docker, you can use a command like the one below. In this example, parameters and sequences from the alphafold dataset are being used and are located at /mnt/alphafold_database on the host machine, and the input files are located in the current working directory. You can adjust the volume mount locations as needed to reflect the locations of your data.
docker run \
--gpus all \
-v $PWD/:/data \
-v /mnt/alphafold_database/:/database \
-ti openfold:latest \
python3 /opt/openfold/run_pretrained_openfold.py \
/data/input.fasta \
/database/pdb_mmcif/mmcif_files/ \
--uniref90_database_path /database/uniref90/uniref90.fasta \
--mgnify_database_path /database/mgnify/mgy_clusters_2018_12.fa \
--pdb70_database_path /database/pdb70/pdb70 \
--uniclust30_database_path /database/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
--output_dir /data \
--bfd_database_path /database/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--model_device cuda:0 \
--jackhmmer_binary_path /opt/conda/bin/jackhmmer \
--hhblits_binary_path /opt/conda/bin/hhblits \
--hhsearch_binary_path /opt/conda/bin/hhsearch \
--kalign_binary_path /opt/conda/bin/kalign \
--param_path /database/params/params_model_1.npzWhile AlphaFold's and, by extension, OpenFold's source code is licensed under
the permissive Apache Licence, Version 2.0, DeepMind's pretrained parameters
fall under the CC BY 4.0 license, a copy of which is downloaded to
openfold/resources/params by the installation script. Note that the latter
replaces the original, more restrictive CC BY-NC 4.0 license as of January 2022.
If you encounter problems using OpenFold, feel free to create an issue! We also welcome pull requests from the community.
For now, cite OpenFold as follows:
@software{Ahdritz_OpenFold_2021,
author = {Ahdritz, Gustaf and Bouatta, Nazim and Kadyan, Sachin and Xia, Qinghui and Gerecke, William and AlQuraishi, Mohammed},
doi = {10.5281/zenodo.5709539},
month = {11},
title = {{OpenFold}},
url = {https://github.com/aqlaboratory/openfold},
year = {2021}
}Any work that cites OpenFold should also cite AlphaFold.