This is an R package that provides support for
3Dmol.js as a
htmlwidgets.
Important: This package is still very early in its development stages. Please give the repository a star on Github if you find the package is useful. In addition, we need your feedback to improve the package, feel free to create an issue if you have any question or feature requirement. You are more than welcome to submit a PR to make any feasible improvements.
You could install the released version of r3dmol from CRAN with:
install.packages("r3dmol")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("swsoyee/r3dmol")library(r3dmol)
r3dmol( # Set up the initial viewer
viewer_spec = m_viewer_spec(
cartoonQuality = 10,
lowerZoomLimit = 50,
upperZoomLimit = 350
)
) %>%
m_add_model( # Add model to scene
data = pdb_6zsl,
format = "pdb"
) %>%
m_zoom_to() %>% # Zoom to encompass the whole scene
m_set_style( # Set style of structures
style = m_style_cartoon(
color = "#00cc96"
)
) %>%
m_set_style( # Set style of specific selection
sel = m_sel(ss = "s"), # (selecting by secondary)
style = m_style_cartoon(
color = "#636efa",
arrows = TRUE
)
) %>%
m_set_style( # Style the alpha helix
sel = m_sel(ss = "h"), # (selecting by alpha helix)
style = m_style_cartoon(
color = "#ff7f0e"
)
) %>%
m_rotate( # Rotate the scene by given angle on given axis
angle = 90,
axis = "y"
) %>%
m_spin() # Animate the scene by spinning it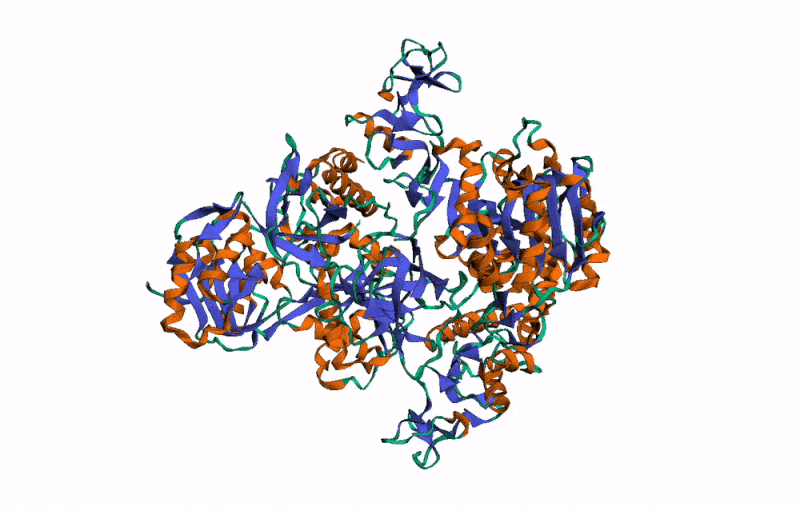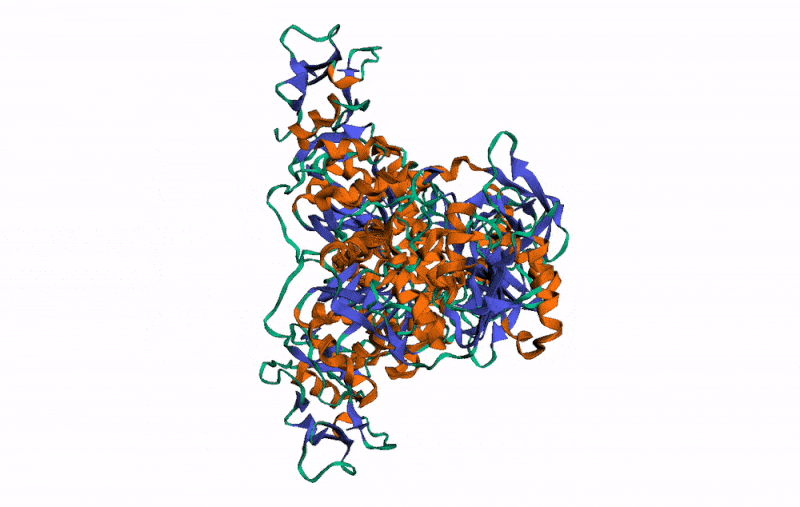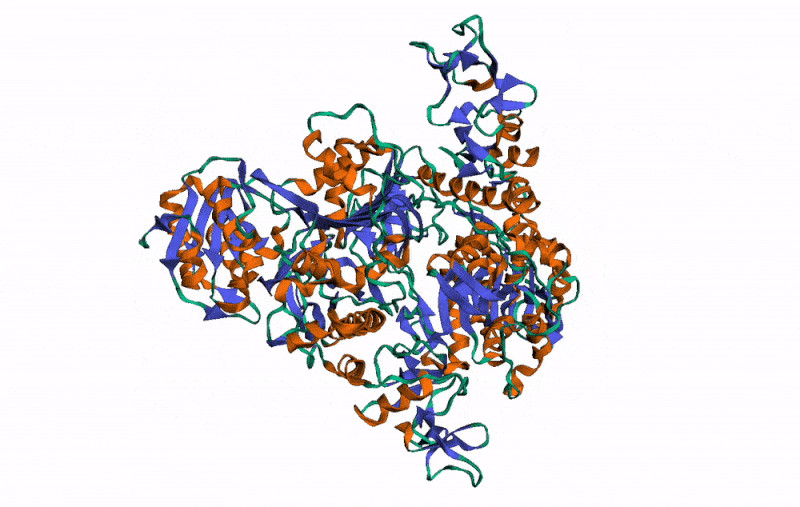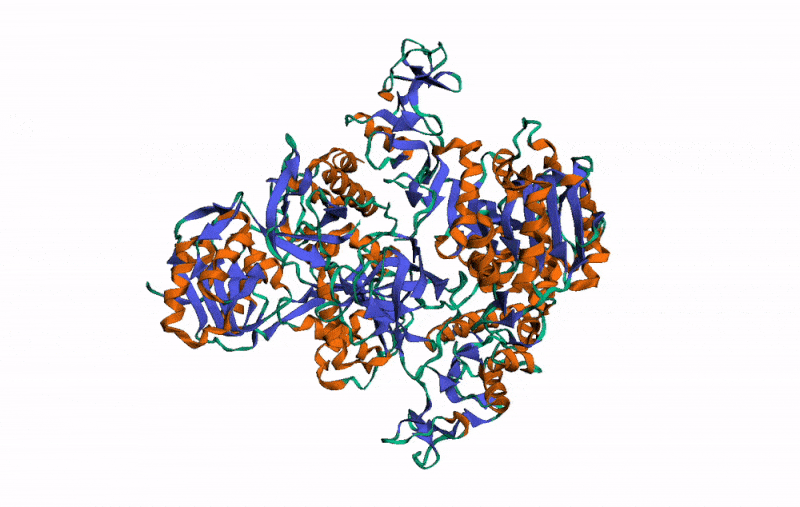
You can find more demo at
here.
Nicholas Rego, David Koes, 3Dmol.js: molecular visualization with WebGL, Bioinformatics, Volume 31, Issue 8, 15 April 2015, Pages 1322–1324,
Please note that the r3dmol project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.

