These models were trained on the dataset https://www.kaggle.com/googleai/pfam-seed-random-split. The task is: given the amino acid sequence of the protein domain, predict which class it belongs. In this case, a subset of the original dataset was utilized.
Multiclass setting, where the protein family must be classified from the raw aminoacid sequence. From the dataset itself, it is important to note two columns:
- sequence: containing the sequence of aminoacids determining the domain, it will be used as input to the model for the classification task.
- family_accession: label that the model will predict in the multi-label classification setting.
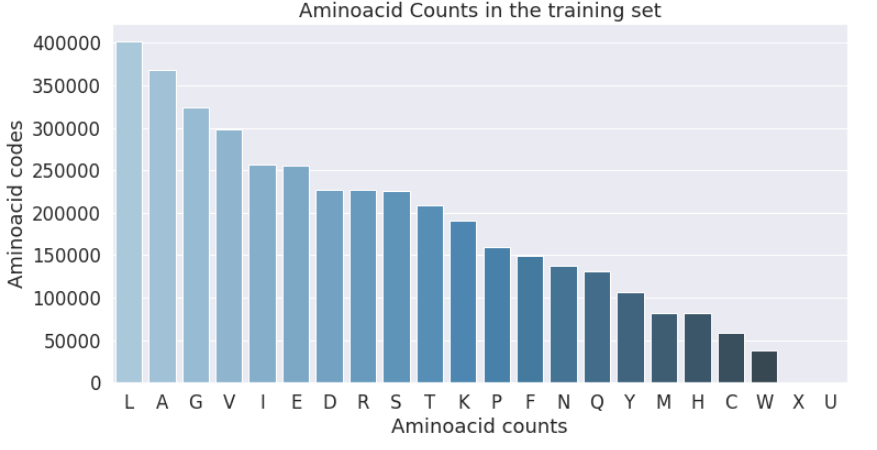
Aminoacids are mapped into a single character. The occurences of some aminoacids are very limited. This is important to notice and usually in NLP applications the least occuring words are mapped into a common token. This is again a design question that I will consider.
- Given the task of multi-class classification, it is necessary to find a model with the right inductive bias given the sequential nature of the input data. For this purpose, it is interesting to read recent papers on NLP, assuming that those methods that perform well for natural language would work in the case of protein sequences.
The first model that I will implement is based on the work of Jennifer M. Johnson et al. (2020) Journal of Chemical Information and Modeling : Deep Dive into Machine Learning for Protein Engineering.
- The proposed model is a convolutional NN with dense layers on top, with the (1-dimensional) convolutional layers would be able to extract relevant features and learn the underlying patterns present in each protein family. Thus, these features extracted by the first layers would be relevant for the classification layers to perform the task.
- Related to inductive biases, it is important to note how this architecture would be shift invariant (translational invariance), thanks to the max pooling layers as well.
Given the success of pre-trained models in NLP, my second solution would be geared towards leveraging transfer learning. In particular, I will be using the model from Burkhard Rost et al. (2020) IEEE Transactions on Pattern Analysis and Machine Intelligence ProtTrans: Towards Cracking the Language of Life’s Code Through Self-Supervised Learning. The model hosted on HuggingFace API ProtBert was trained on a self-supervised task, namely MLM (Masked-Language Model).
- After downloading the model with the trained parameters, I will add additional layers for the classifier and then fine-tune the whole resulting model in our downstream task.
- Pre-training on a MLM objective also allows to learn an internal representation which can be used by the classification layers to incorporate useful features. Transformer-based models capitlize on the attention mechanism, such that a certain element of the sequence can attend on another element, thus in a sense it is able to generate contextual embeddings.
It is also important to note how the paper which produced the dataset by Bileschi et al. (2019) BioArxiv Can Deep Learning Classify the Protein Universe is based on a 1-D CNN model as well.
-
CNN-based model is able to achieve 97% accuracy on the held-out set, thus proving that a convolutional model is able to capture the dependencies and underlying patterns in the protein sequence. Using an embedding layer and batch normalization provide the right inductive bias for learning representations of the protein sequences. Also, running the model on a GPU allows for an extremely fast learning with the subset of the data that I was using.
-
The fine-tuned transformer model is able to achieve 99% accuracy on the test set, showcasing the power of transfer learning for downstream tasks. The main drawback would be both training time and inference time, which would require a lot more computational power. This implementation allows further interpretability of the model by visualizing attention weights in the transformer architecture.
-
It is also remarkable how both models are robust with respect to short sequences and truncation set up to only 128 characters as well.
Further work:
- Test the same models trained on the whole dataset, given access to more computational power.
- Another interesting point would be related to few-shot learning. Siamese networks have shown to perform well without many labelled examples. More precisely, it would be interesting to leverage the same transformer architecture to obtain the learned representation and compute similarity metrics among samples to perform the classification. This type of siamese transformer architectures have been shown to perform well in NLP in the work by Shuai Gao et al. (2021) TBSN: Sparse-Transformer Based Siamese Network for Few-Shot Action Recognition.