scVelo is a scalable toolkit for estimating and analyzing RNA velocities in single cells using dynamical modeling.
The methods used herein are based on our preprint Bergen et al. (2019).
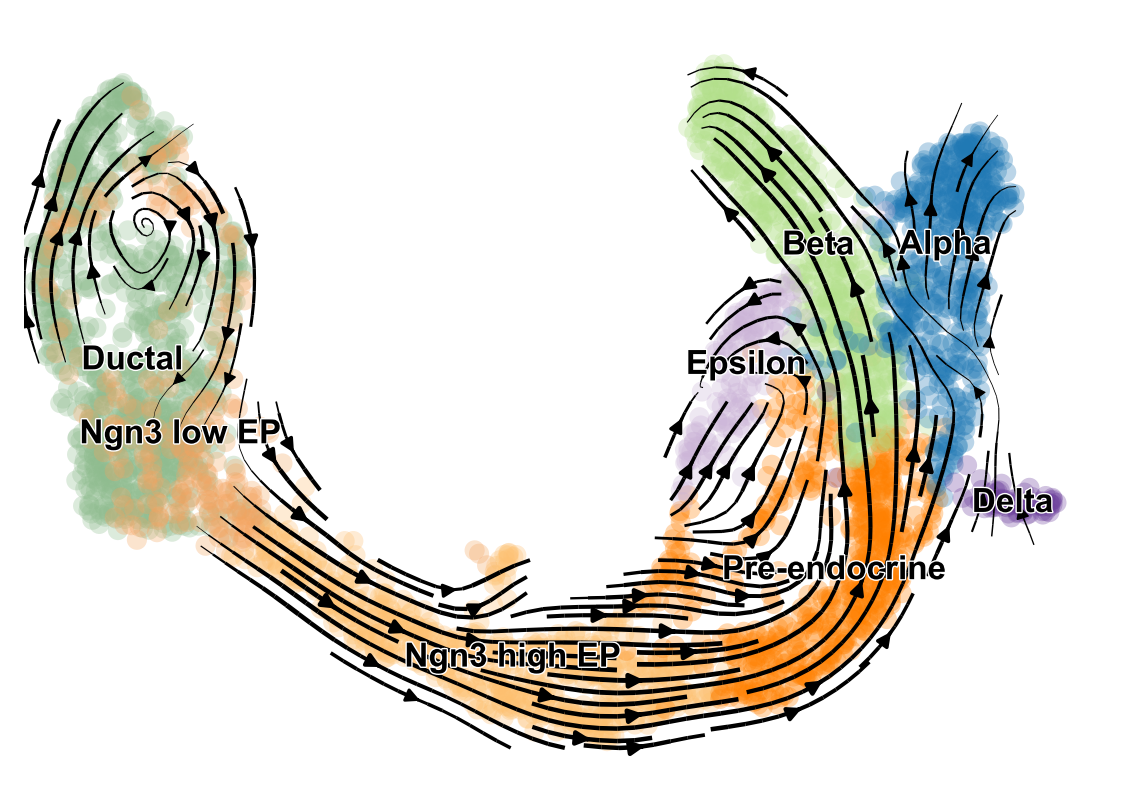
RNA velocity, the time derivative of mRNA abundance, enables you to infer directionality in your data by superimposing splicing information. The main principles have been presented in La Manno et al. (2018), and are based on a deterministic steady-state model of transcriptional dynamics. scVelo provides two extensions as described in Bergen et al. (2019): A stochastic model that incorporates second-order moments, and a dynamical model that captures the full splicing kinetics. It thereby adapts RNA velocity to widely varying specifications such as non-stationary populations.
It is compatible with scanpy (Wolf et al., 2018). Making use of sparse implementation, iterative neighbors search and other techniques, it is remarkably efficient in terms of memory and runtime without loss in accuracy and runs easily on your local machine (30k cells in a few minutes).
Install from PyPI:
pip install -U scvelo
See the the documentation at https://scvelo.org for details, which includes:
- How scVelo estimates RNA velocity: https://scvelo.org/about.html
- How to install and getting started: https://scvelo.org/getting_started.html
- About the API: https://scvelo.org/api.html
- Tutorials: http://tutorials.scvelo.org/Pancreas.html
Your feedback, in particular any issue you stumble upon, is highly appreciated and addressed to [email protected].