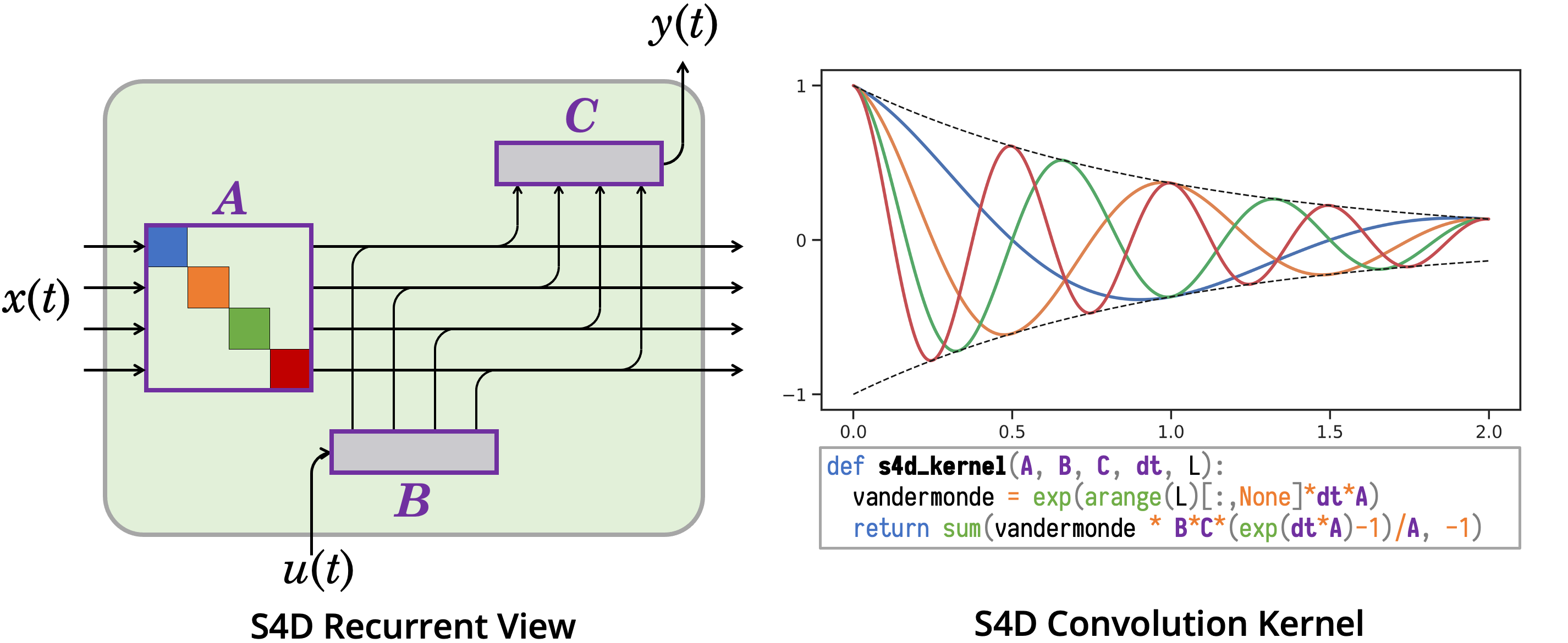
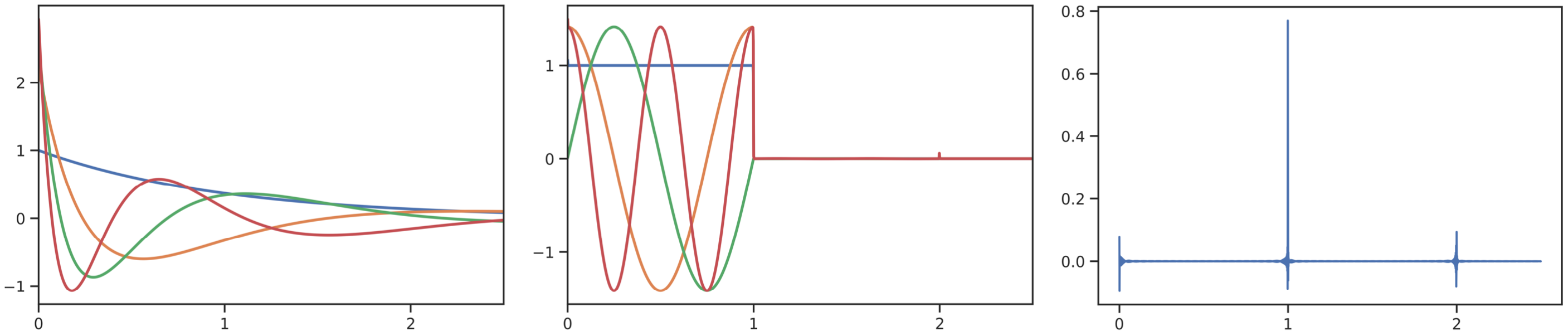
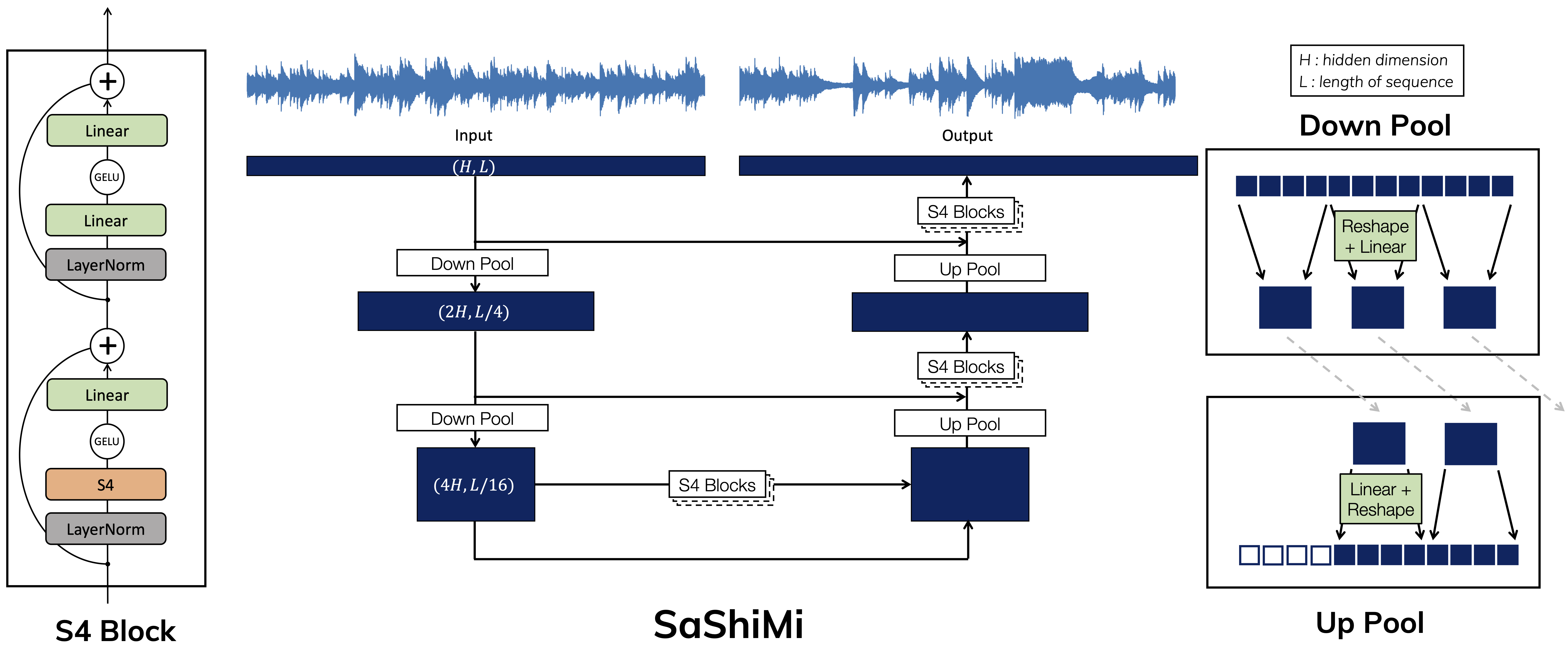
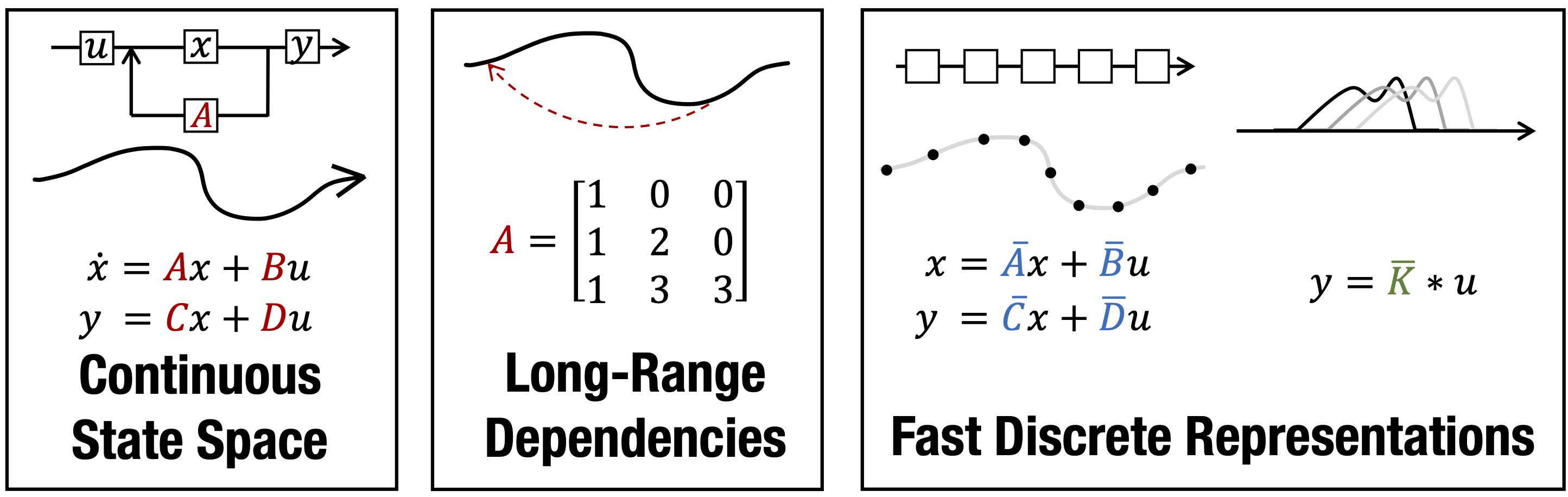
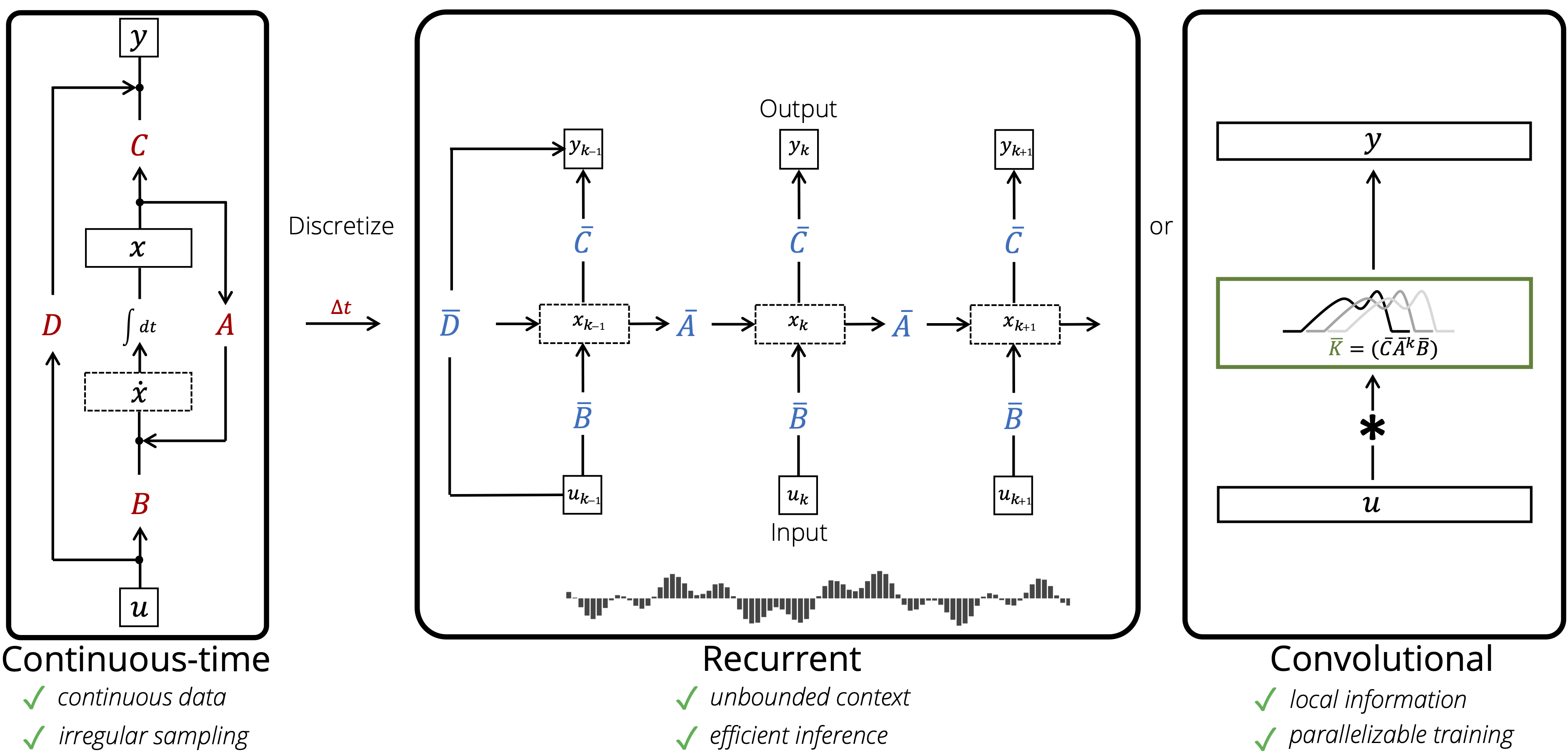
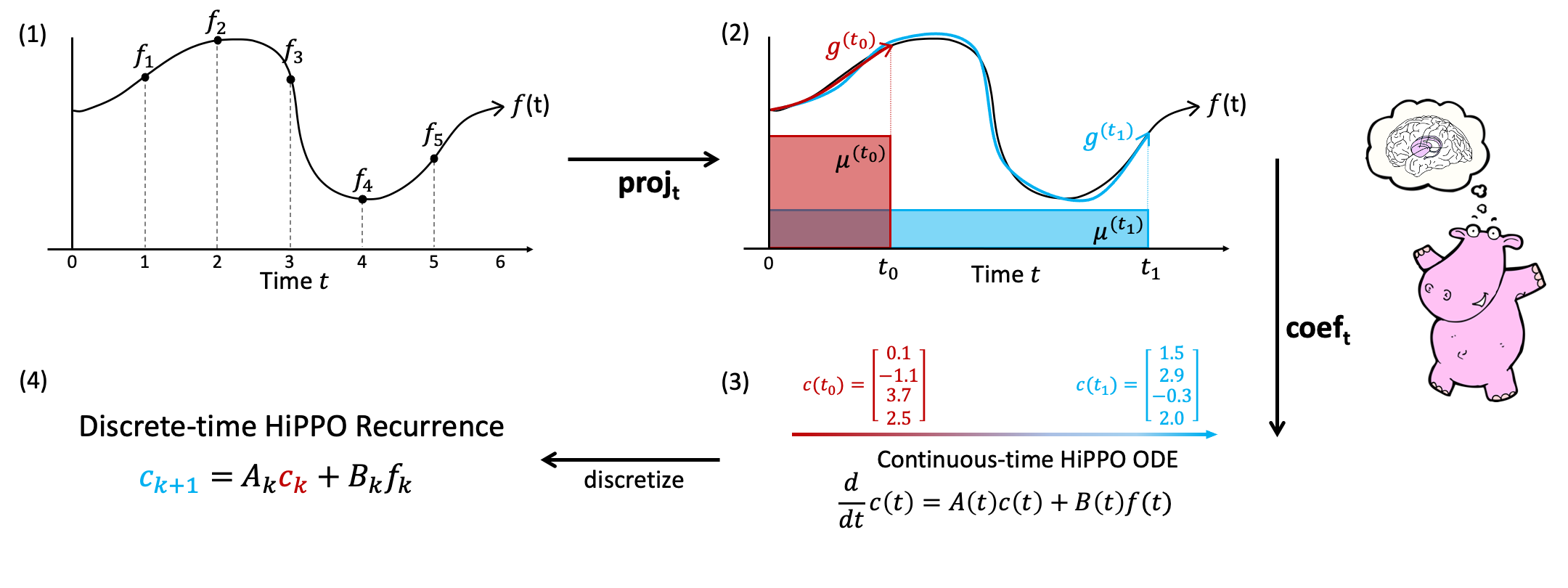
This repository provides implementations and experiments for the following papers.
On the Parameterization and Initialization of Diagonal State Space Models
Albert Gu, Ankit Gupta, Karan Goel, Christopher Ré
Paper: https://arxiv.org/abs/2206.11893
Other variants including DSS and GSS are also supported. DSS is the predecessor to S4D that is also available in its own fork.
How to Train Your HiPPO: State Spaces with Generalized Orthogonal Basis Projections
Albert Gu*, Isys Johnson*, Aman Timalsina, Atri Rudra, Christopher Ré
Paper: https://arxiv.org/abs/2206.12037
It's Raw! Audio Generation with State-Space Models
Karan Goel, Albert Gu, Chris Donahue, Christopher Ré
Paper: https://arxiv.org/abs/2202.09729
Efficiently Modeling Long Sequences with Structured State Spaces
Albert Gu, Karan Goel, Christopher Ré
Paper: https://arxiv.org/abs/2111.00396
Combining Recurrent, Convolutional, and Continuous-time Models with the Linear State Space Layer
Albert Gu, Isys Johnson, Karan Goel, Khaled Saab, Tri Dao, Atri Rudra, Christopher Ré
Paper: https://arxiv.org/abs/2110.13985
HiPPO: Recurrent Memory with Optimal Polynomial Projections
Albert Gu*, Tri Dao*, Stefano Ermon, Atri Rudra, Christopher Ré
Paper: https://arxiv.org/abs/2008.07669
Setting up the environment and porting S4 to external codebases:
Reproducing experiments from the papers:
Using this repository for training models:
See CHANGELOG.md
- More documentation for training from scratch using this repository
- Compilation of S4 resources and implementations
- pip package
This repository requires Python 3.8+ and Pytorch 1.10+. Other packages are listed in requirements.txt.
A core operation of S4 is the "Cauchy kernel" described in the paper.
This is actually a very simple operation; a naive implementation of this operation can be found in the standalone in the function cauchy_naive.
However, as the paper describes, this has suboptimal memory usage that currently requires a custom kernel to overcome in PyTorch.
Two more efficient methods are supported. The code will automatically detect if either of these is installed and call the appropriate kernel.
This version is faster but requires manual compilation for each machine environment.
Run python setup.py install from the directory extensions/cauchy/.
This version is provided by the pykeops library.
Installation usually works out of the box with pip install pykeops cmake which are also listed in the requirements file.
Self-contained files for the S4 layer and variants can be found in src/models/s4/, which includes instructions for calling the module.
See notebooks/ for visualizations explaining some concepts behind HiPPO and S4.
example.py is a self-contained training script for MNIST and CIFAR that imports the standalone S4 file. The default settings python example.py reaches 88% accuracy on sequential CIFAR with a very simple S4D model of 200k parameters.
This script can be used as an example for using S4 in external repositories.
This repository aims to provide a very flexible framework for training sequence models. Many models and datasets are supported.
Basic usage is python -m train, or equivalently
python -m train pipeline=mnist model=s4
which trains an S4 model on the Permuted MNIST dataset. This should get to around 90% after 1 epoch which takes 1-3 minutes depending on GPU.
More examples of using this repository can be found in Experiments and Training.
One important feature of this codebase is supporting parameters that require different optimizer hyperparameters.
In particular, the SSM kernel is particularly sensitive to the
See the method register in the model (e.g. s4d.py) and the function setup_optimizer in the training script (e.g. example.py) for an examples of how to implement this in external repos.
Figures from the HTTYH and S4D papers can be visualized from notebooks/. These include animations of HiPPO and S4 that were used in various S4 talks. The animation code can also be found in a .py file instead of notebook.
Instructions for reproducing experiments from the papers can be found in experiments.md.
Basic datasets are auto-downloaded, including MNIST, CIFAR, and Speech Commands. All logic for creating and loading datasets is in src/dataloaders directory. The README inside this subdirectory documents how to download and organize other datasets.
Models are defined in src/models. See the README in this subdirectory for an overview.
The core training infrastructure of this repository is based on Pytorch-Lightning with a configuration scheme based on Hydra.
The main entrypoint is train.py and configs are found in configs/.
Pre-defined configs for many end-to-end experiments are provided (see experiments.md).
Configs can also be easily modified through the command line. An example experiment is
python -m train pipeline=mnist dataset.permute=True model=s4 model.n_layers=3 model.d_model=128 model.norm=batch model.prenorm=True wandb=null
This uses the Permuted MNIST task with an S4 model with a specified number of layers, backbone dimension, and normalization type.
See configs/README.md for more detailed documentation about the configs.
It is recommended to read the Hydra documentation to fully understand the configuration framework. For help launching specific experiments, please file an issue.
Each experiment will be logged to its own directory (generated by Hydra) of the form ./outputs/<date>/<time>/. Checkpoints will be saved here inside this folder and printed to console whenever a new checkpoint is created.
To resume training, simply point to the desired .ckpt file (a PyTorch Lightning checkpoint, e.g. ./outputs/<date>/<time>/checkpoints/val/loss.ckpt) and append the flag train.ckpt=<path>/<to>/<checkpoint>.ckpt to the original training command.
The PTL Trainer class controls the overall training loop and also provides many useful pre-defined flags. Some useful examples are explained below. The full list of allowable flags can be found in the PTL documentation, as well as our trainer configs. See the default trainer config configs/trainer/default.yaml for the most useful options.
Simply pass in trainer.gpus=2 to train with 2 GPUs.
trainer.weights_summary=full prints out every layer of the model with their parameter counts. Useful for debugging internals of models.
trainer.limit_{train,val}_batches={10,0.1} trains (validates) on only 10 batches (0.1 fraction of all batches). Useful for testing the train loop without going through all the data.
Logging with WandB is built into this repository.
In order to use this, simply set your WANDB_API_KEY environment variable, and change the wandb.project attribute of configs/config.yaml (or pass it on the command line e.g. python -m train .... wandb.project=s4).
Set wandb=null to turn off WandB logging.
Autoregressive generation can be performed with the generate.py script. This script can be used in two ways after training a model using this codebase.
The more flexible option requires the checkpoint path of the trained PyTorch Lightning model.
The generation script accepts the same config options as the train script, with a few additional flags that are documented in configs/generate.yaml.
After training with python -m train <train flags>, generate with
python -m generate <train flags> checkpoint_path=<path/to/model.ckpt> <generation flags>
Any of the flags found in the config can be overridden.
Note: This option can be used with either .ckpt checkpoints (PyTorch Lightning, which includes information for the Trainer) or .pt checkpoints (PyTorch, which is just a model state dict).
The second option for generation does not require passing in training flags again, and instead reads the config from the Hydra experiment folder, along with a PyTorch Lightning checkpoint within the experiment folder.
Download the WikiText-103 model checkpoint, for example to ./checkpoints/s4-wt103.pt.
This model was trained with the command python -m train experiment=lm/s4-wt103. Note that from the config we can see that the model was trained with a receptive field of length 8192.
To generate, run
python -m generate experiment=lm/s4-wt103 checkpoint_path=checkpoints/s4-wt103.pt n_samples=1 l_sample=16384 l_prefix=8192 decode=text
This generates a sample of length 16384 conditioned on a prefix of length 8192.
Let's train a small SaShiMi model on the SC09 dataset. We can also reduce the number of training and validation batches to get a checkpoint faster:
python -m train experiment=audio/sashimi-sc09 model.n_layers=2 trainer.limit_train_batches=0.1 trainer.limit_val_batches=0.1
After the first epoch completes, a message is printed indicating where the checkpoint is saved.
Epoch 0, global step 96: val/loss reached 3.71754 (best 3.71754), saving model to "<repository>/outputs/<date>/<time>/checkpoints/val/loss.ckpt"
Option 1:
python -m generate experiment=audio/sashimi-sc09 model.n_layers=2 checkpoint_path=<repository>/outputs/<date>/<time>/checkpoints/val/loss.ckpt n_samples=4 l_sample=16000
This option redefines the full config so that the model and dataset can be constructed.
Option 2:
python -m generate experiment_path=<repository>/outputs/<date>/<time> checkpoint_path=checkpoints/val/loss.ckpt n_samples=4 l_sample=16000
This option only needs the path to the Hydra experiment folder and the desired checkpoint within.
configs/ config files for model, data pipeline, training loop, etc.
data/ default location of raw data
extensions/ CUDA extension for Cauchy kernel
src/ main source code for models, datasets, etc.
callbacks/ training loop utilities (e.g. checkpointing)
dataloaders/ dataset and dataloader definitions
models/ model definitions
tasks/ encoder/decoder modules to interface between data and model backbone
utils/
sashimi/ SaShiMi README and additional code (generation, metrics, MTurk)
example.py Example training script for using S4 externally
train.py Training entrypoint for this repo
generate.py Autoregressive generation script
In addition to this top level README, several READMEs detailing the usage of this repository are organized in subdirectories.
- src/dataloaders/README.md
- src/models/README.md
- src/models/s4/README.md
- experiments.md
- configs/README.md
- configs/model/README.md
- configs/experiment/README.md
- sashimi/README.md
If you use this codebase, or otherwise found our work valuable, please cite:
@article{gu2022s4d,
title={On the Parameterization and Initialization of Diagonal State Space Models},
author={Gu, Albert and Gupta, Ankit and Goel, Karan and R\'e, Christopher},
journal={arXiv preprint arXiv:2206.11893},
year={2022}
}
@article{gu2022hippo,
title={How to Train Your HiPPO: State Space Models with Generalized Basis Projections},
author={Gu, Albert and Johnson, Isys and Timalsina, Aman and Rudra, Atri and R\'e, Christopher},
journal={arXiv preprint arXiv:2206.12037},
year={2022}
}
@article{goel2022sashimi,
title={It's Raw! Audio Generation with State-Space Models},
author={Goel, Karan and Gu, Albert and Donahue, Chris and R{\'e}, Christopher},
journal={International Conference on Machine Learning ({ICML})},
year={2022}
}
@inproceedings{gu2022efficiently,
title={Efficiently Modeling Long Sequences with Structured State Spaces},
author={Gu, Albert and Goel, Karan and R\'e, Christopher},
booktitle={The International Conference on Learning Representations ({ICLR})},
year={2022}
}
@article{gu2021combining,
title={Combining Recurrent, Convolutional, and Continuous-time Models with Linear State-Space Layers},
author={Gu, Albert and Johnson, Isys and Goel, Karan and Saab, Khaled and Dao, Tri and Rudra, Atri and R{\'e}, Christopher},
journal={Advances in neural information processing systems},
volume={34},
year={2021}
}
@article{gu2020hippo,
title={HiPPO: Recurrent Memory with Optimal Polynomial Projections},
author={Gu, Albert and Dao, Tri and Ermon, Stefano and Rudra, Atri and R{\'e}, Christopher},
journal={Advances in neural information processing systems},
volume={33},
year={2020}
}