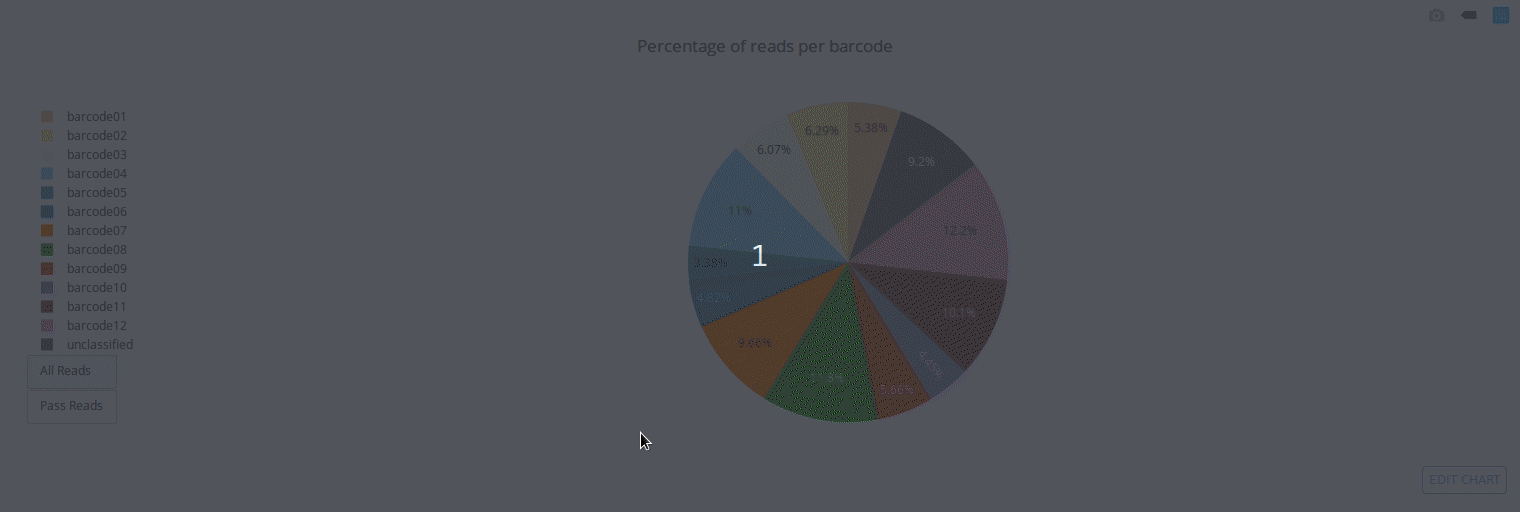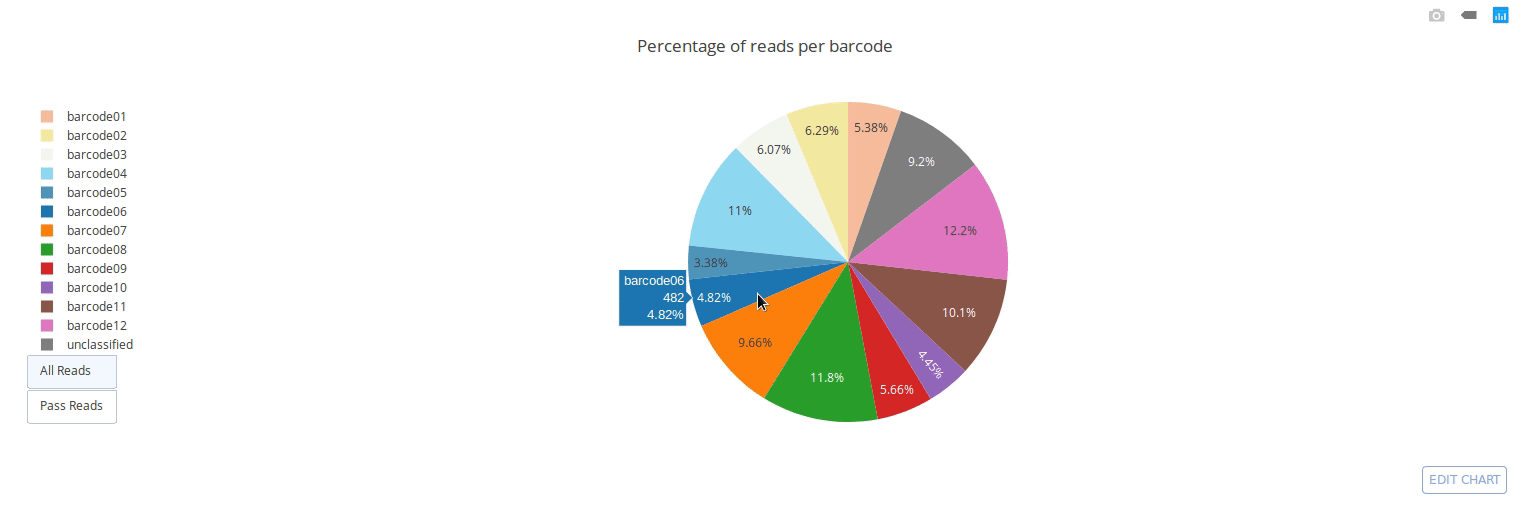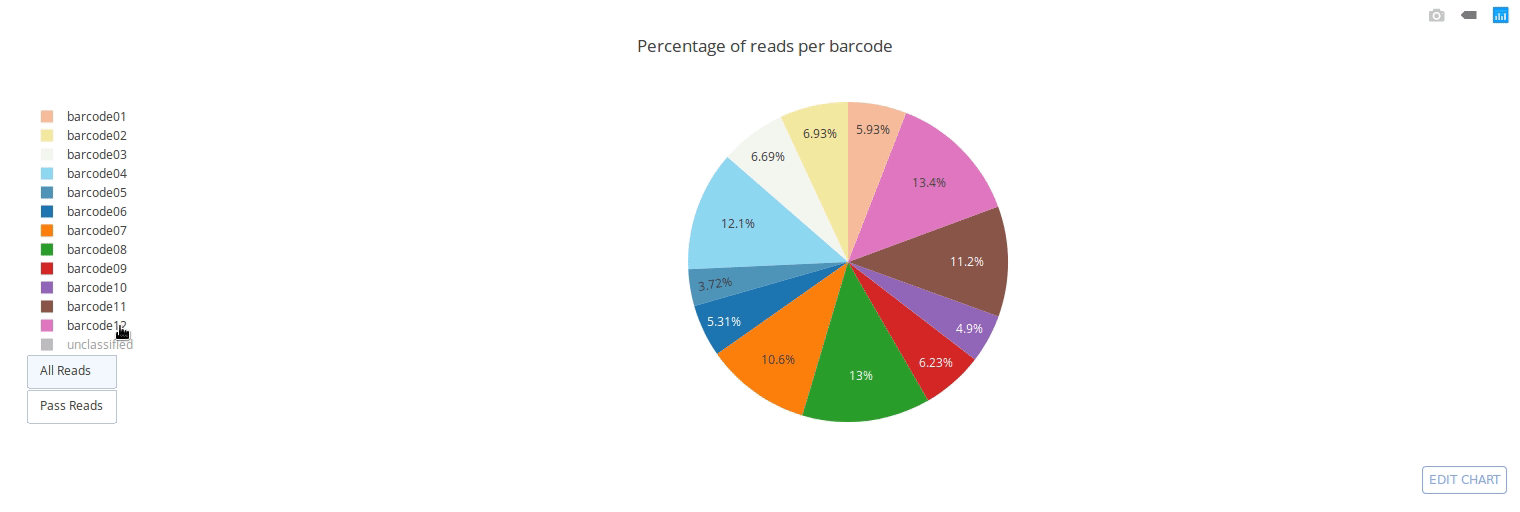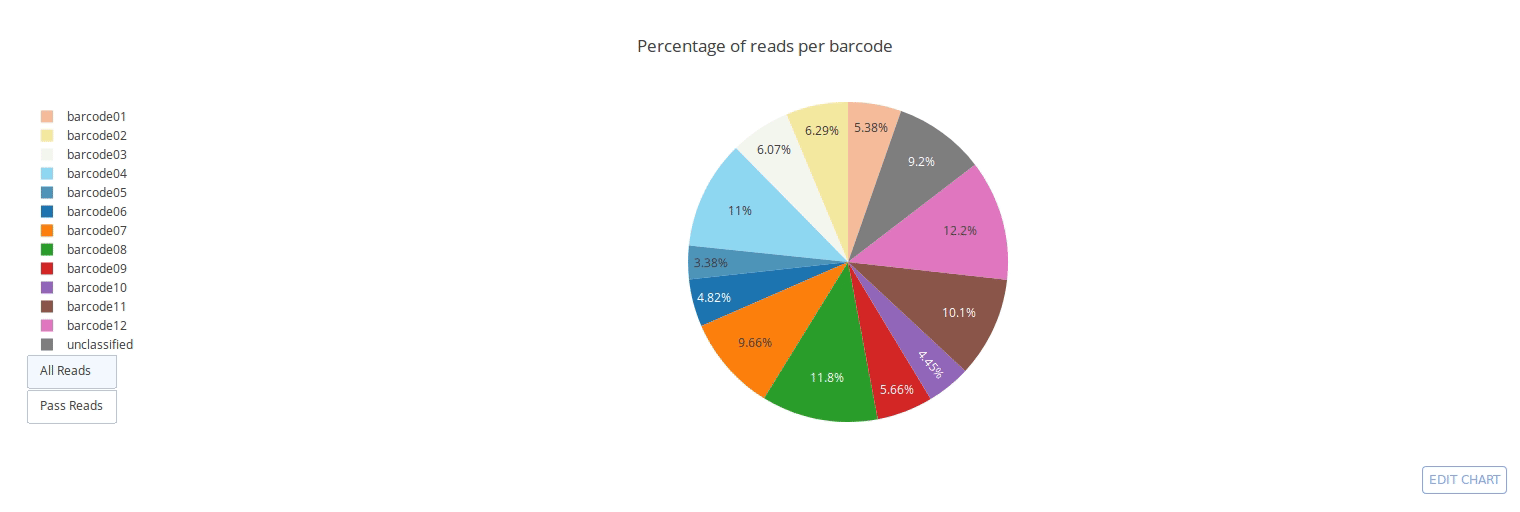
Full documentation is available at https://a-slide.github.io/pycoQC/
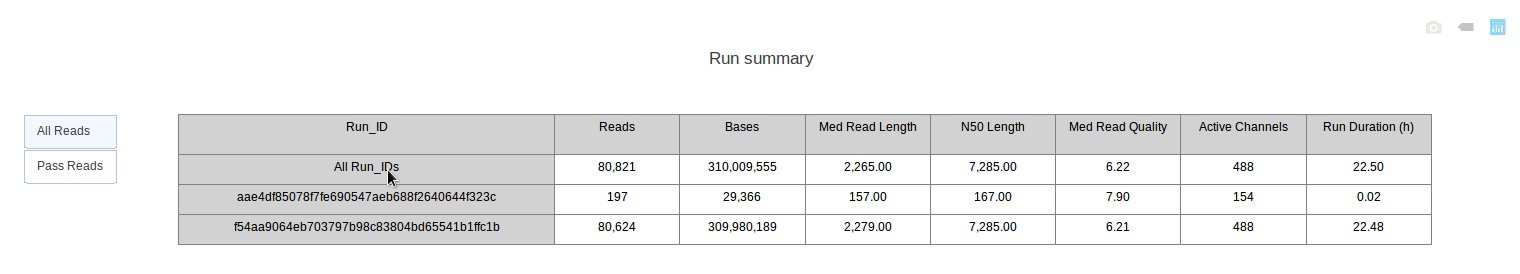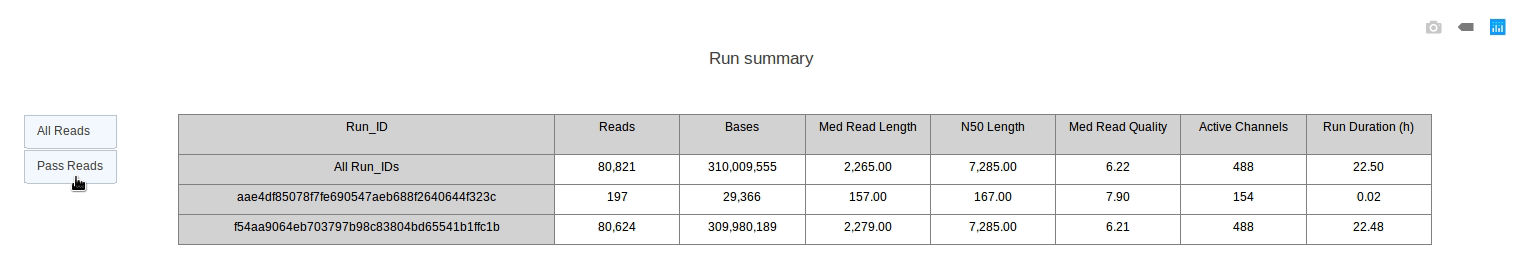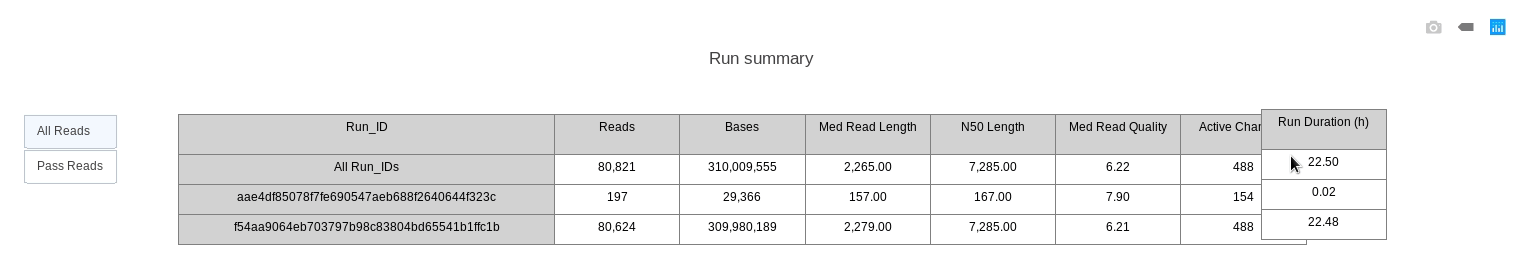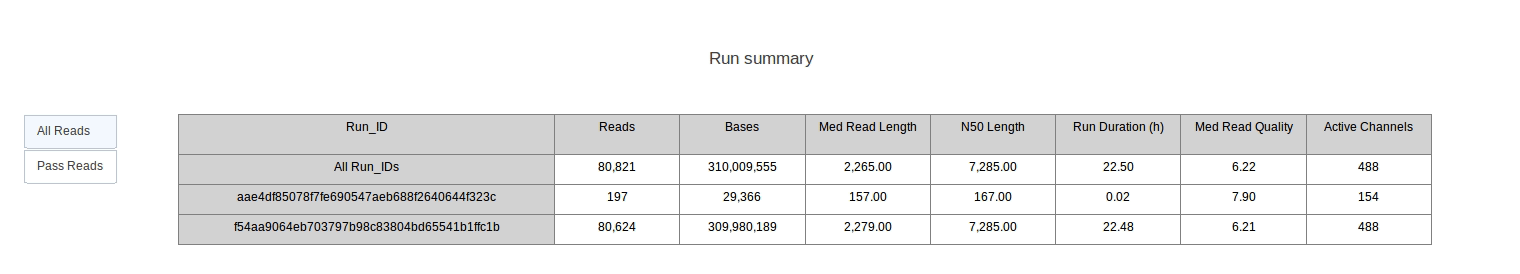
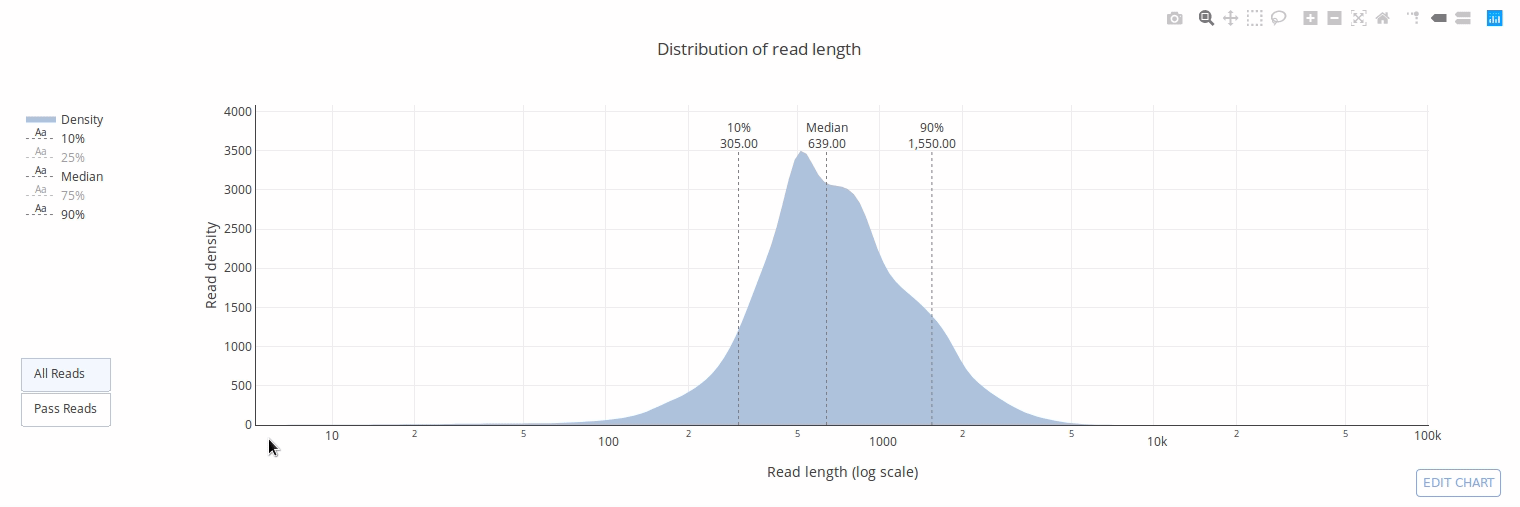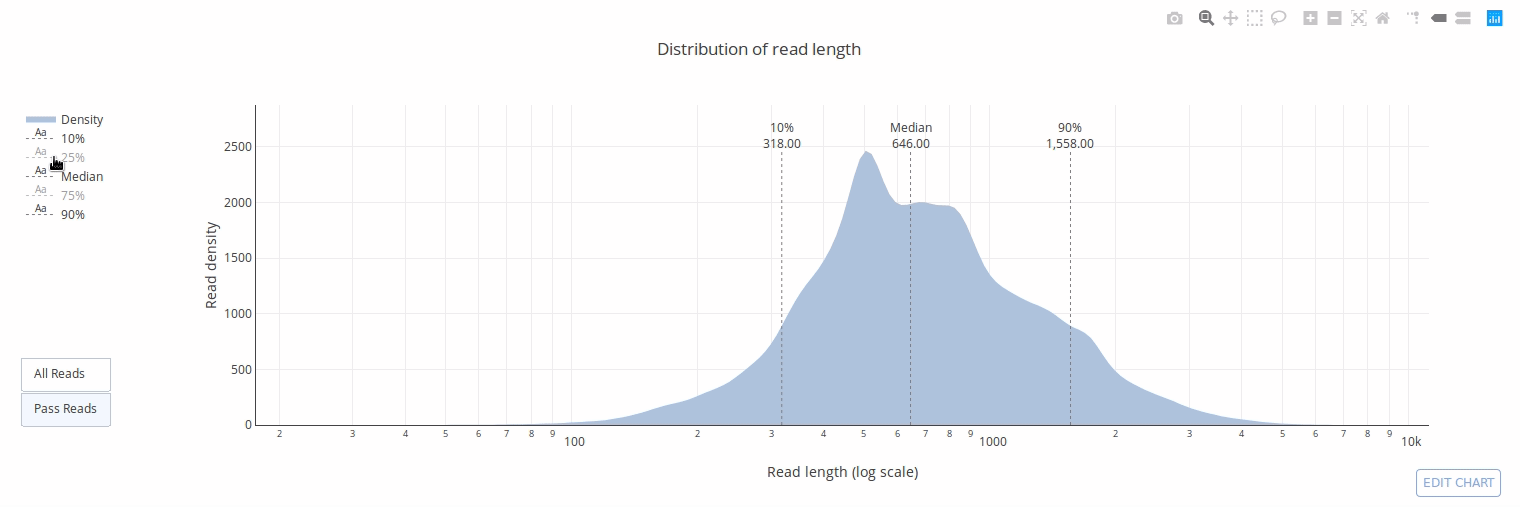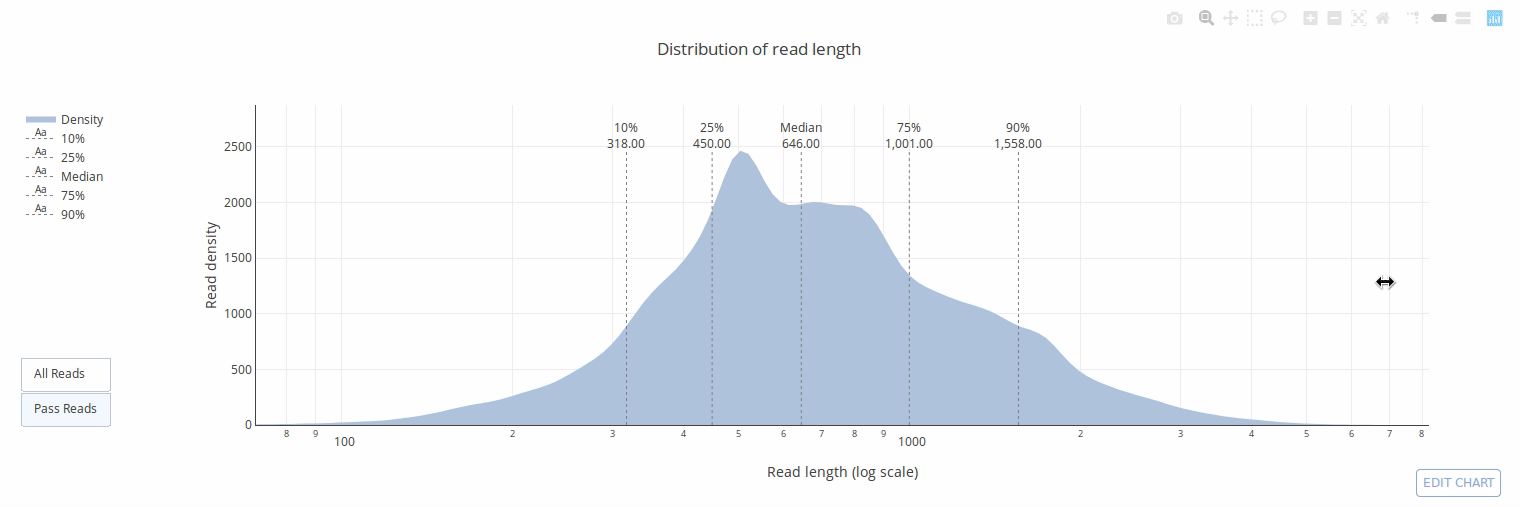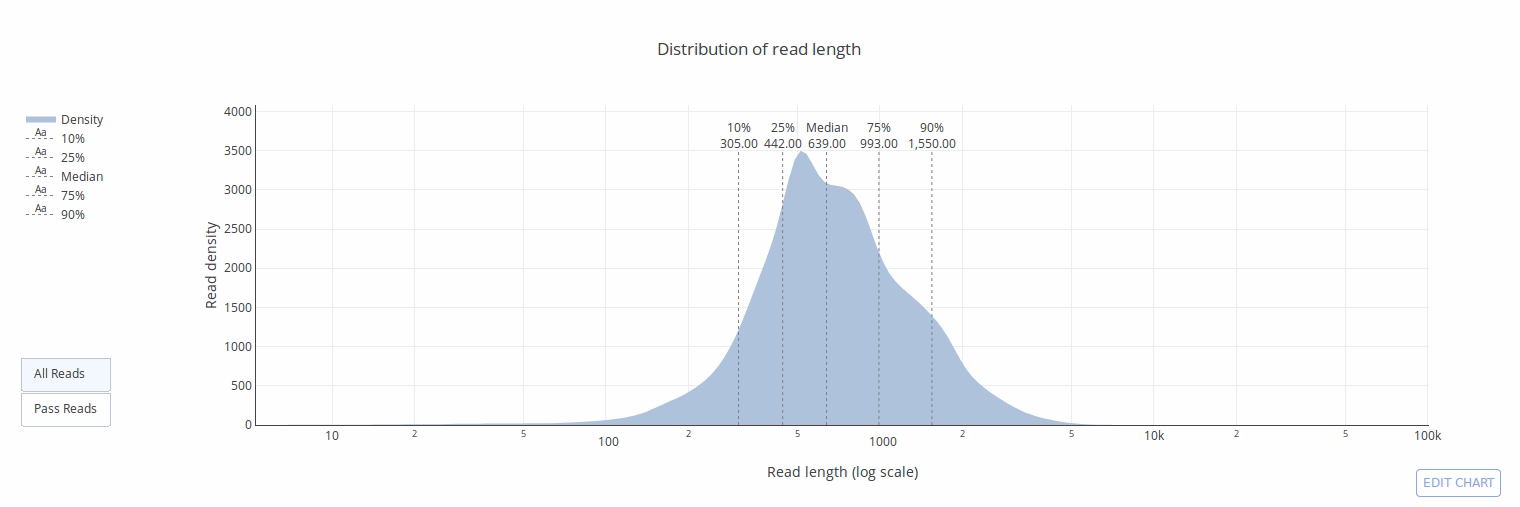
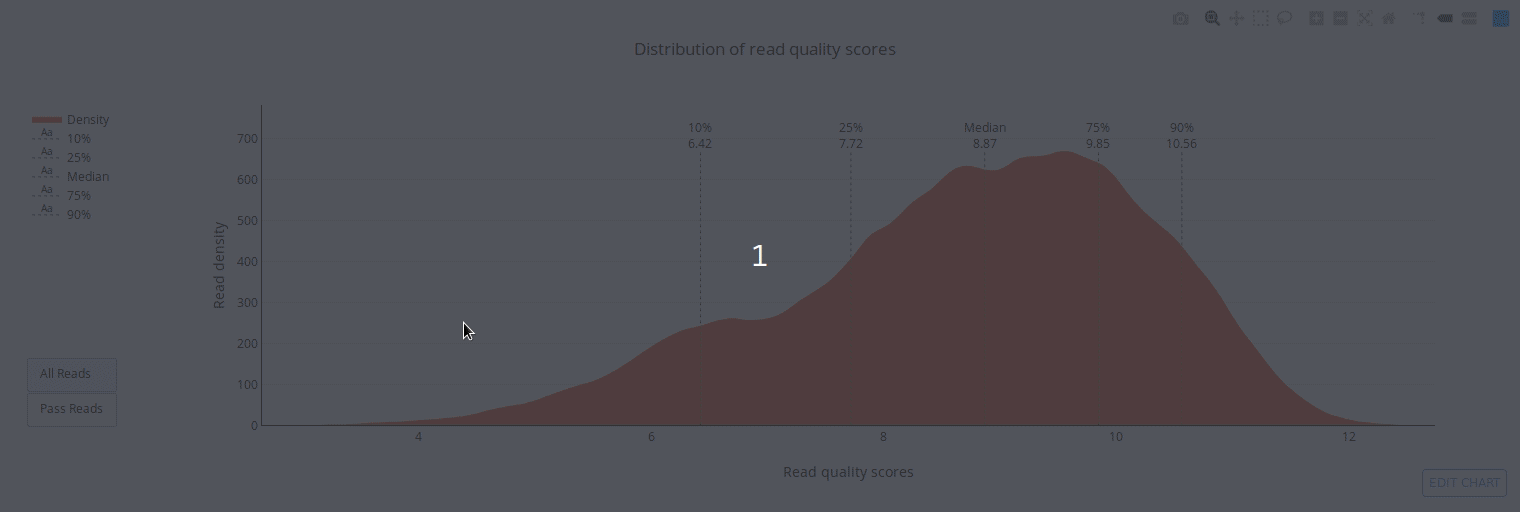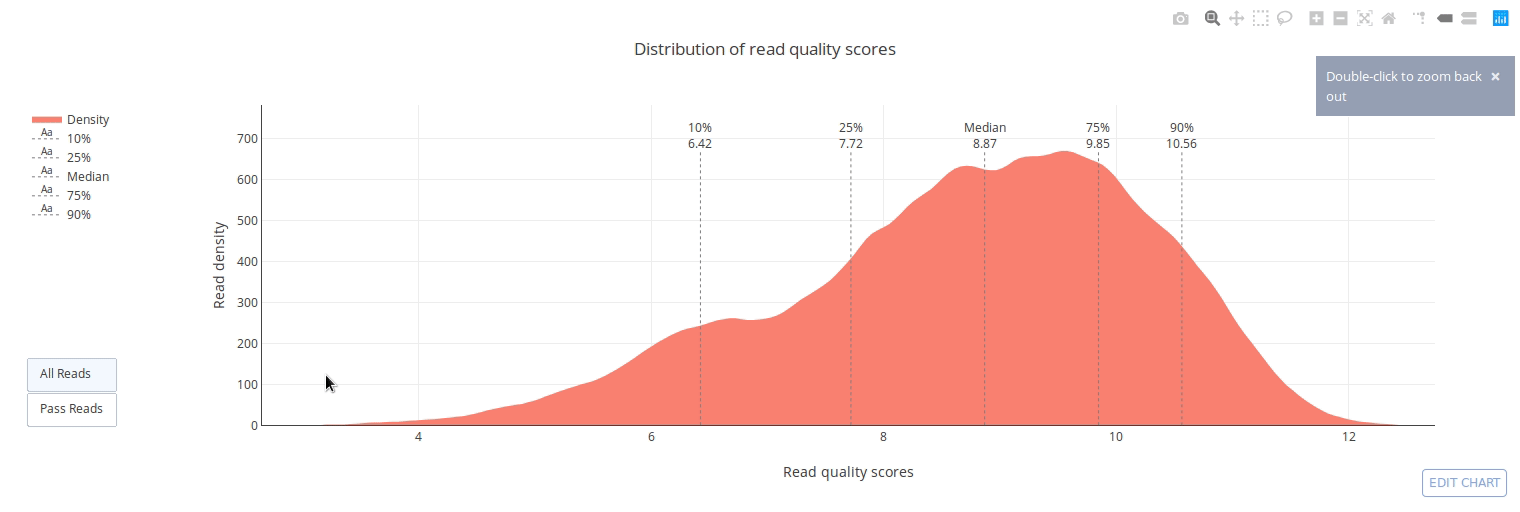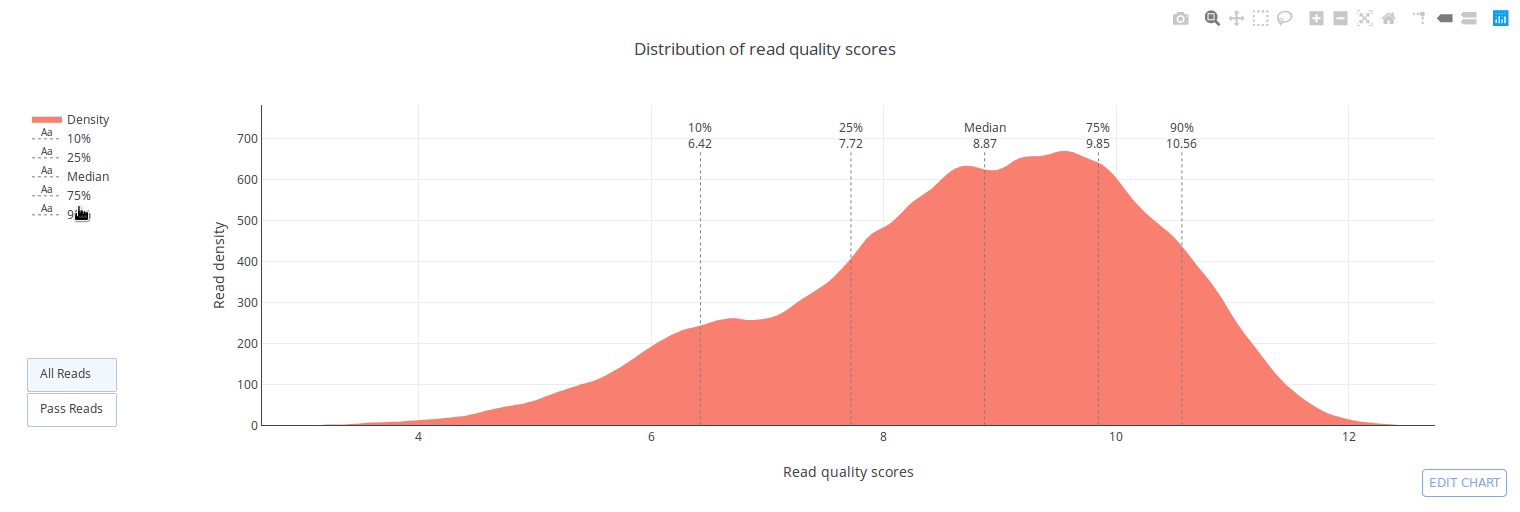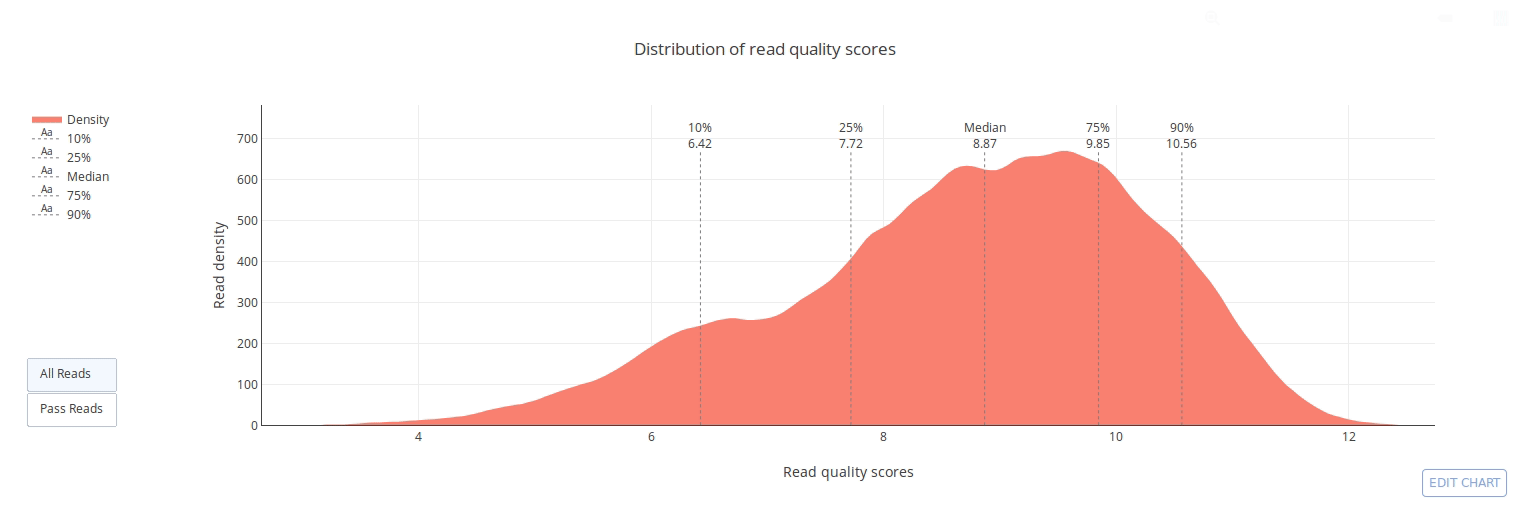
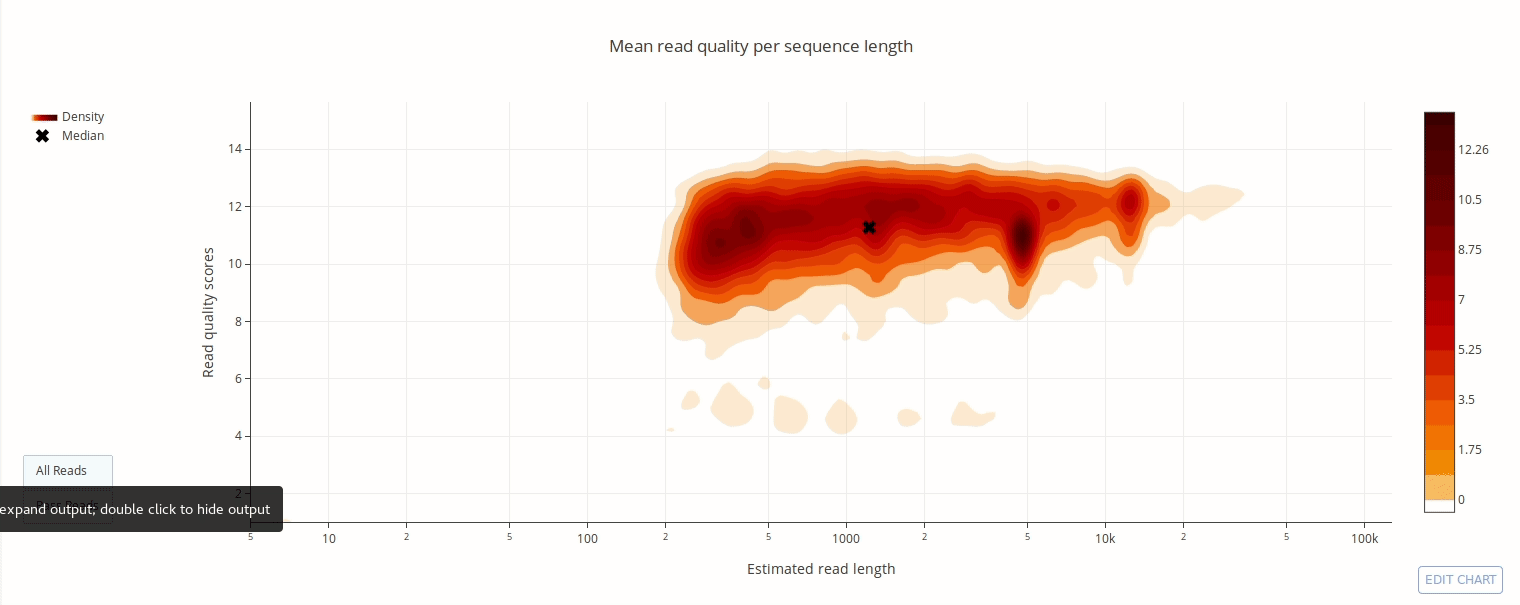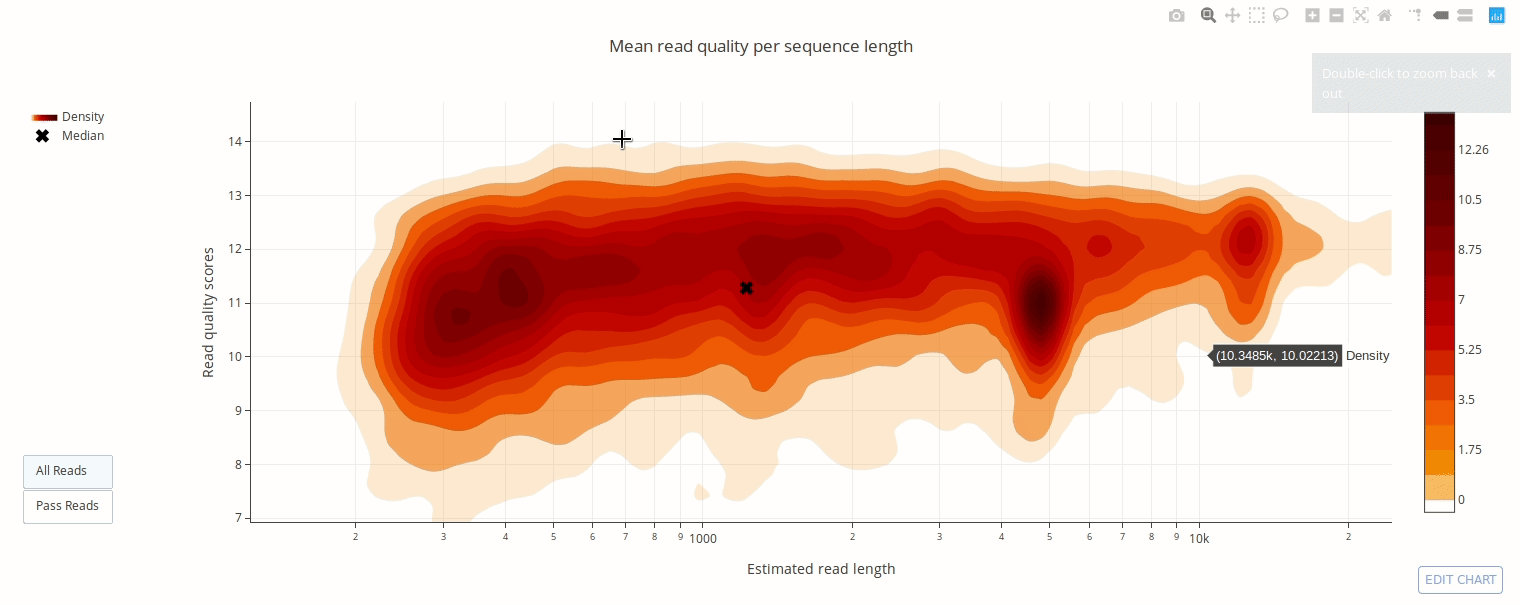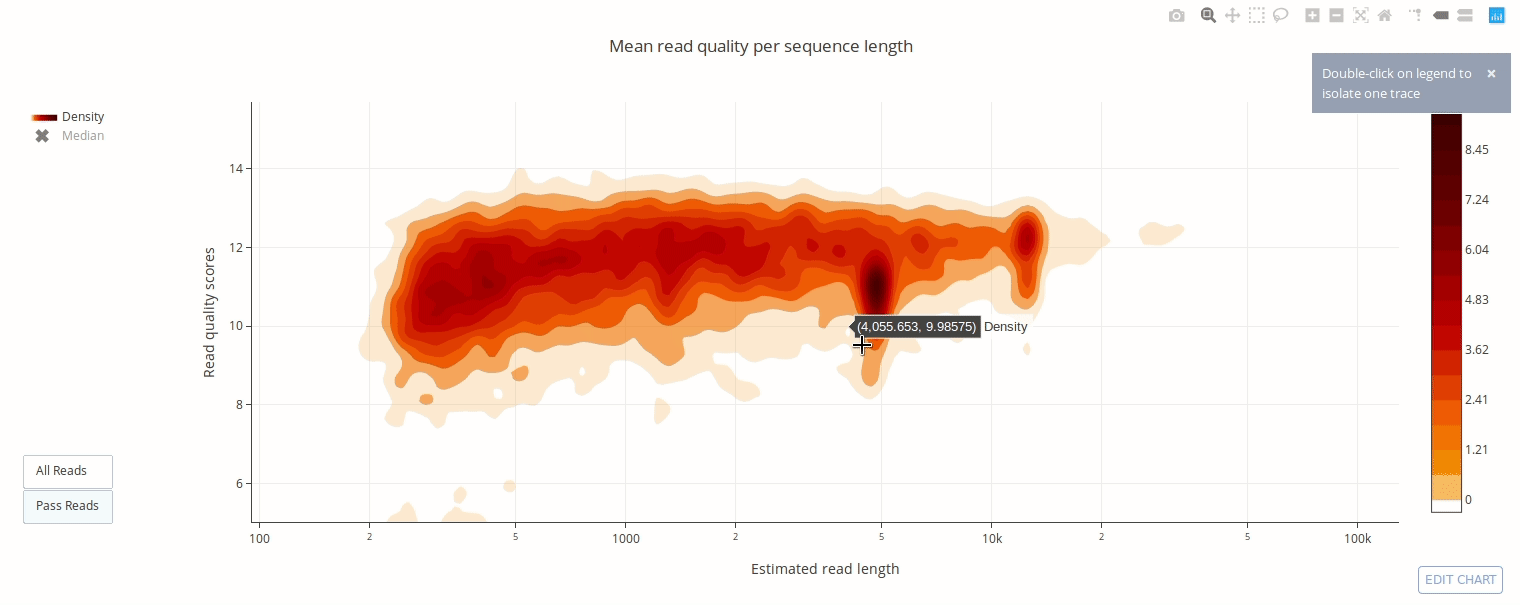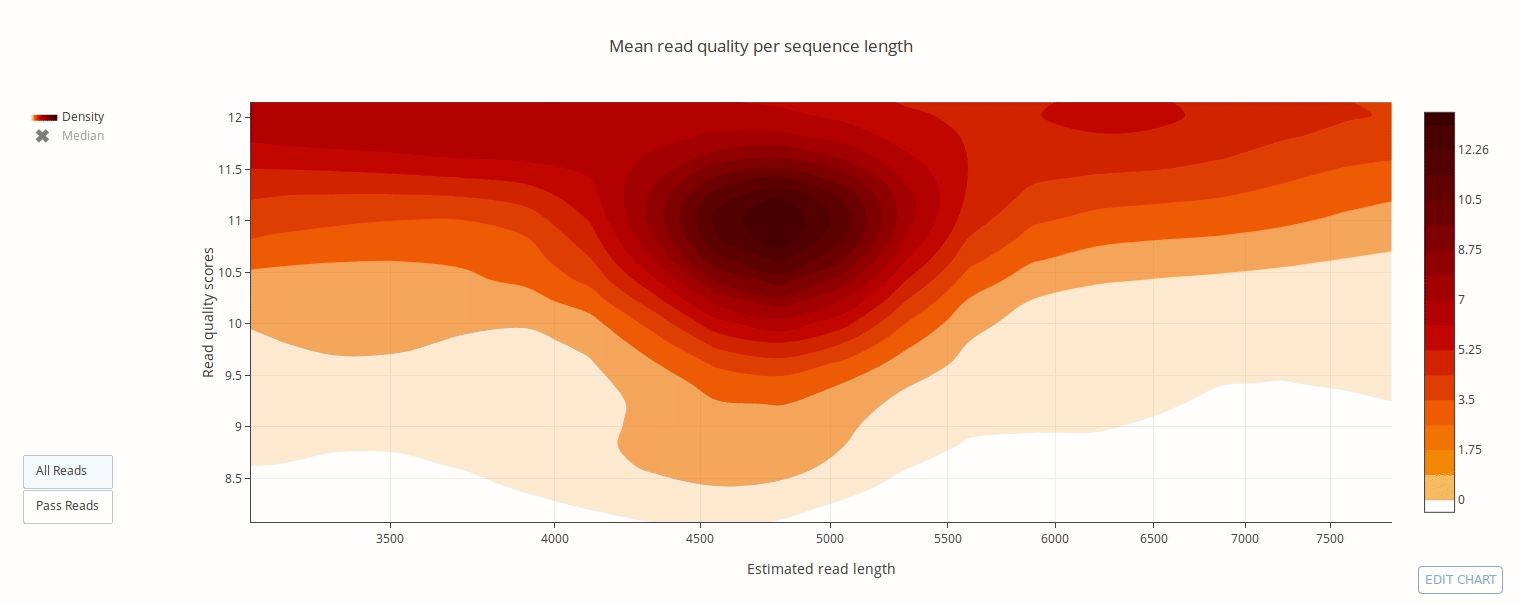
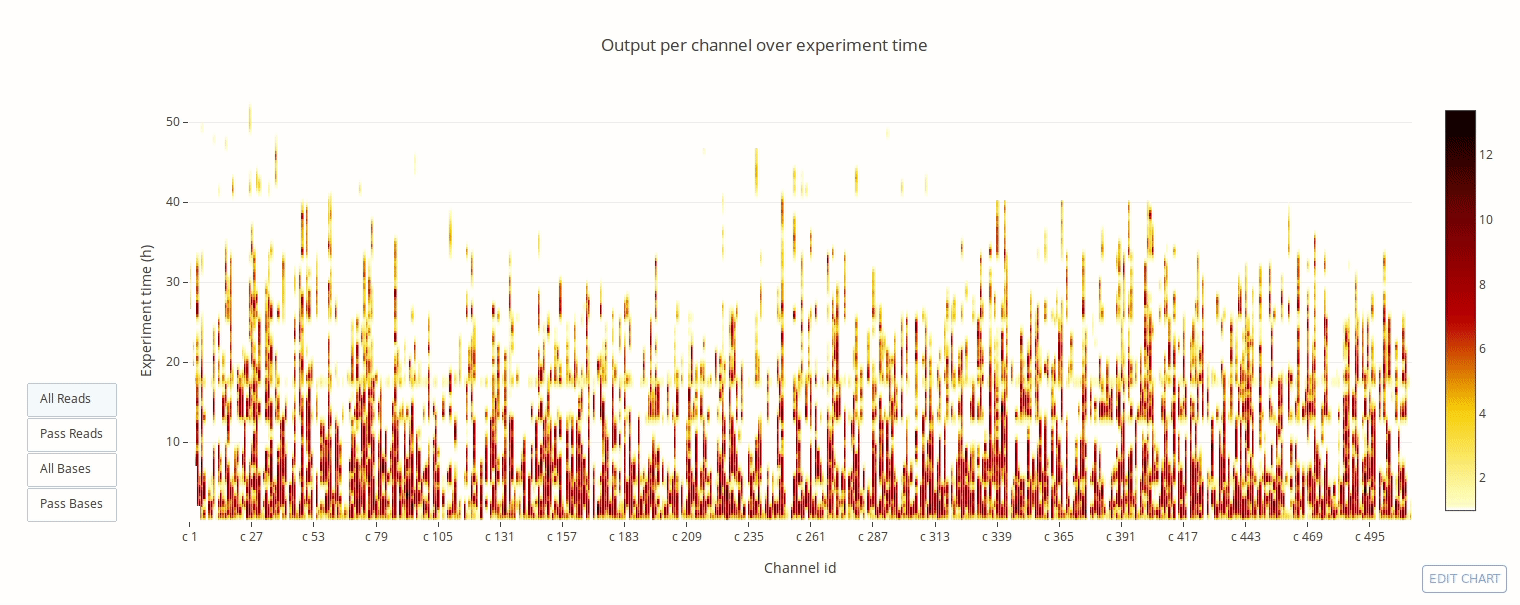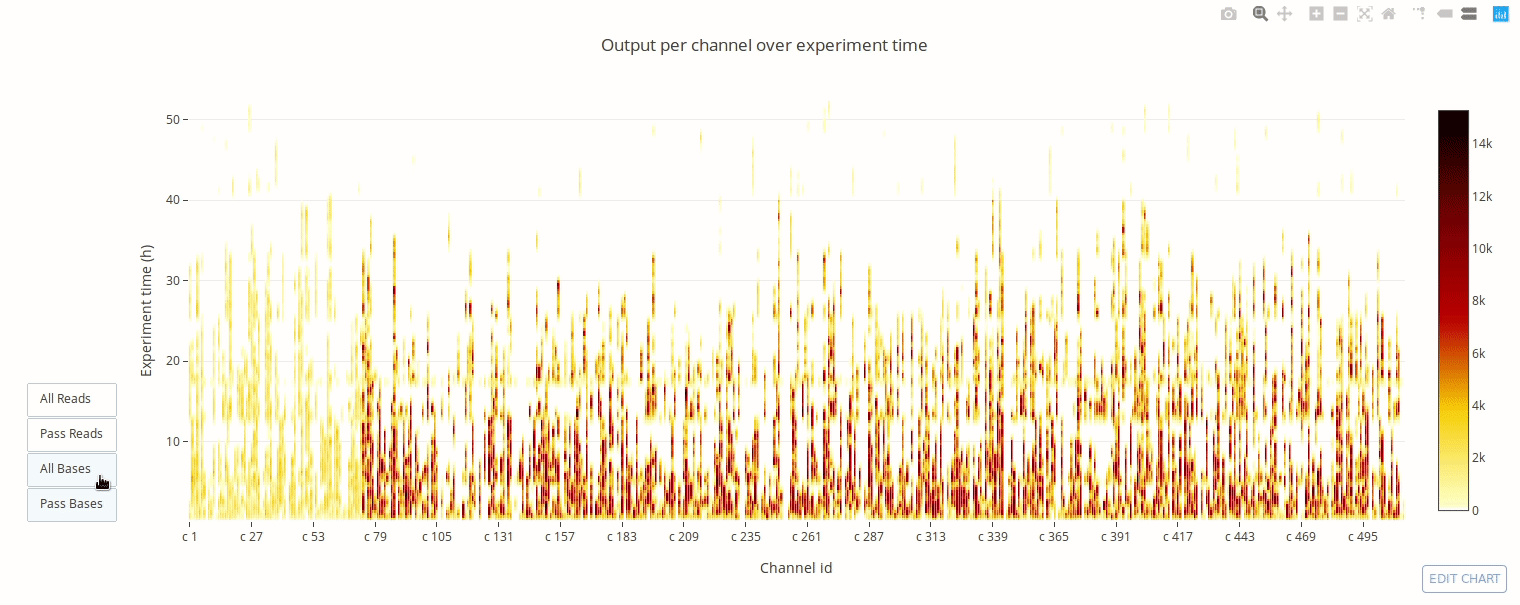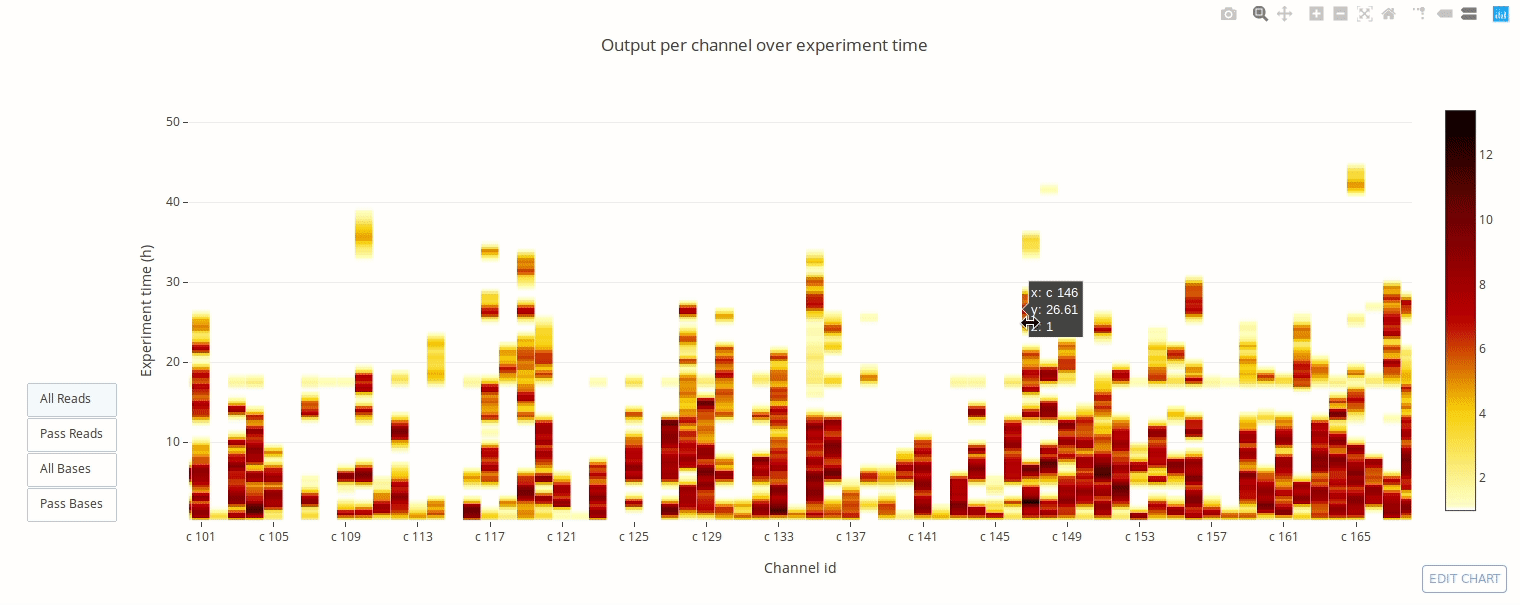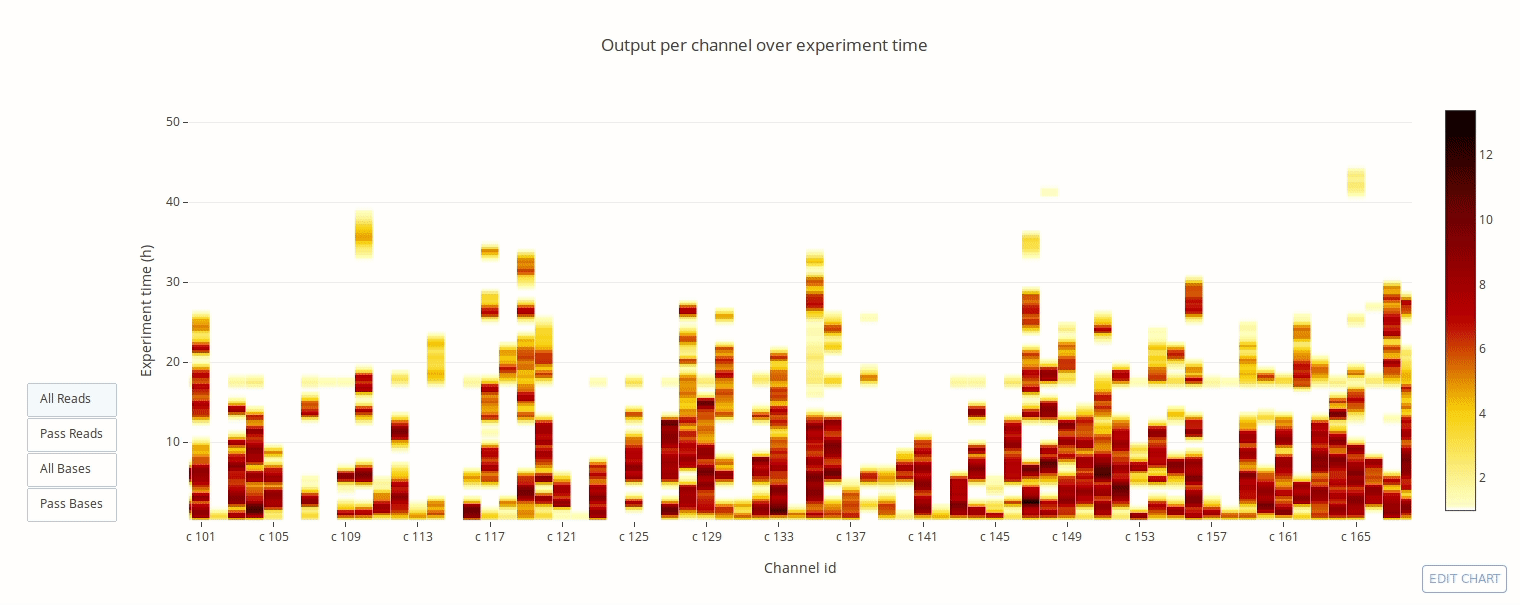
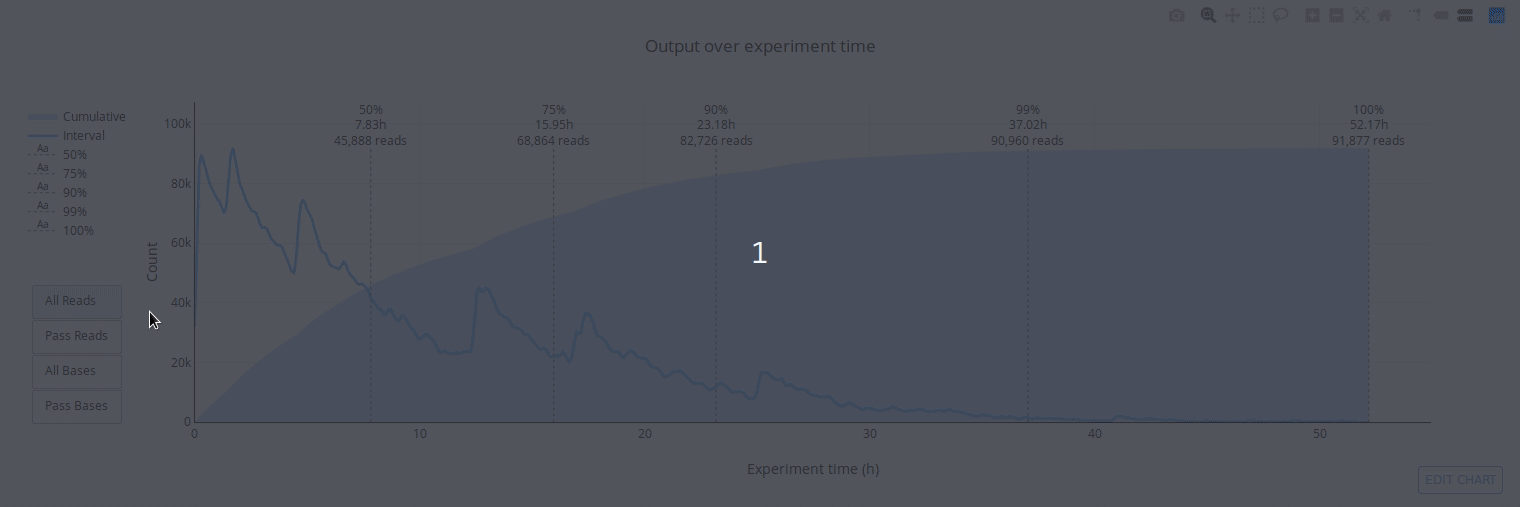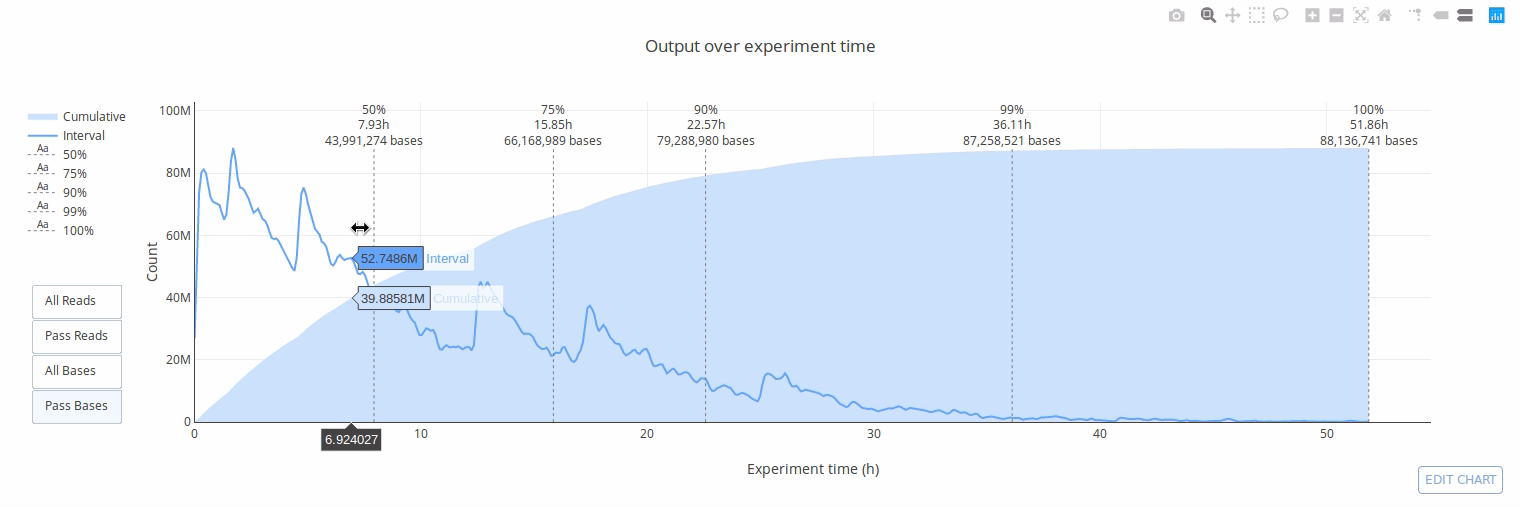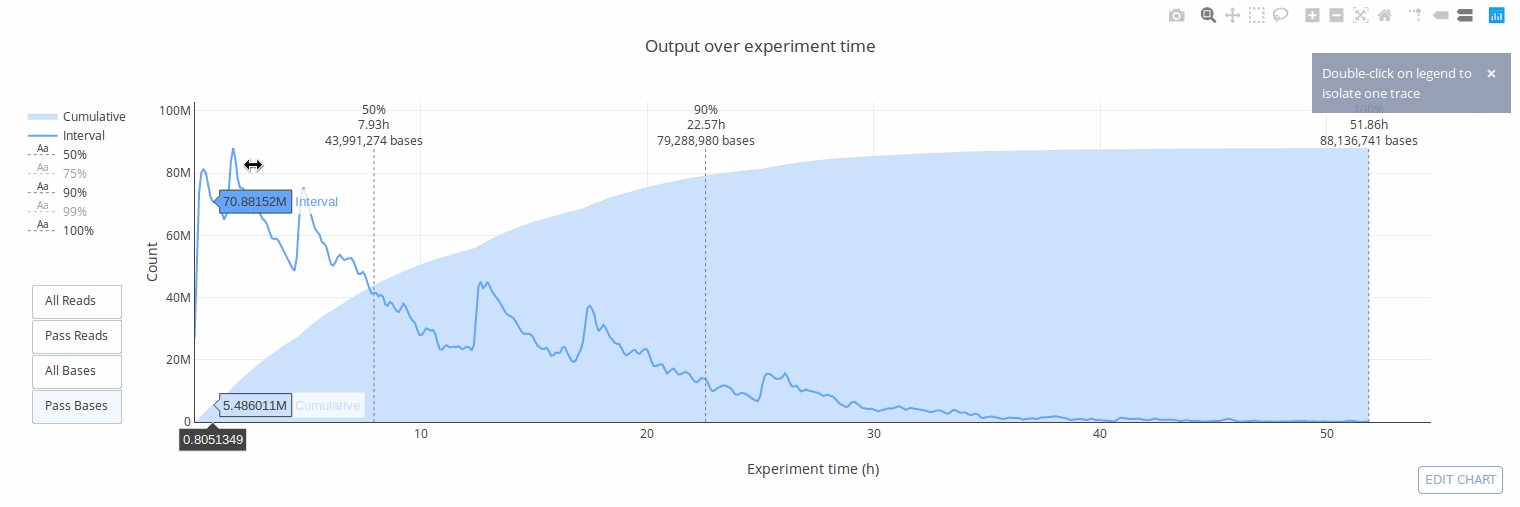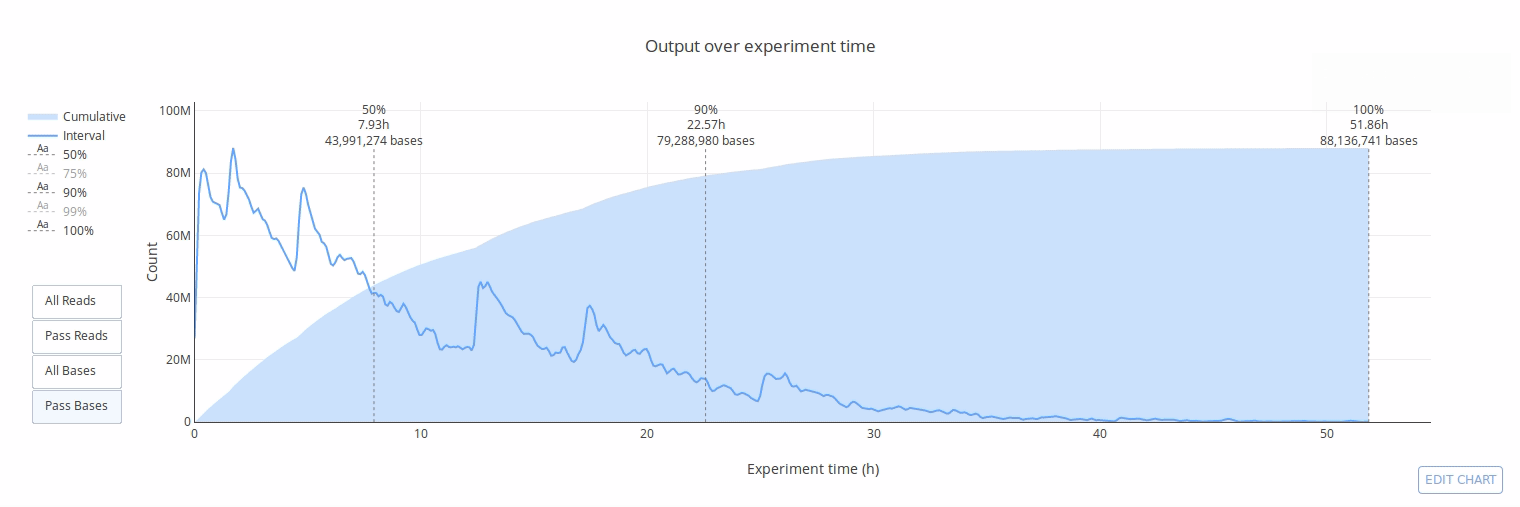
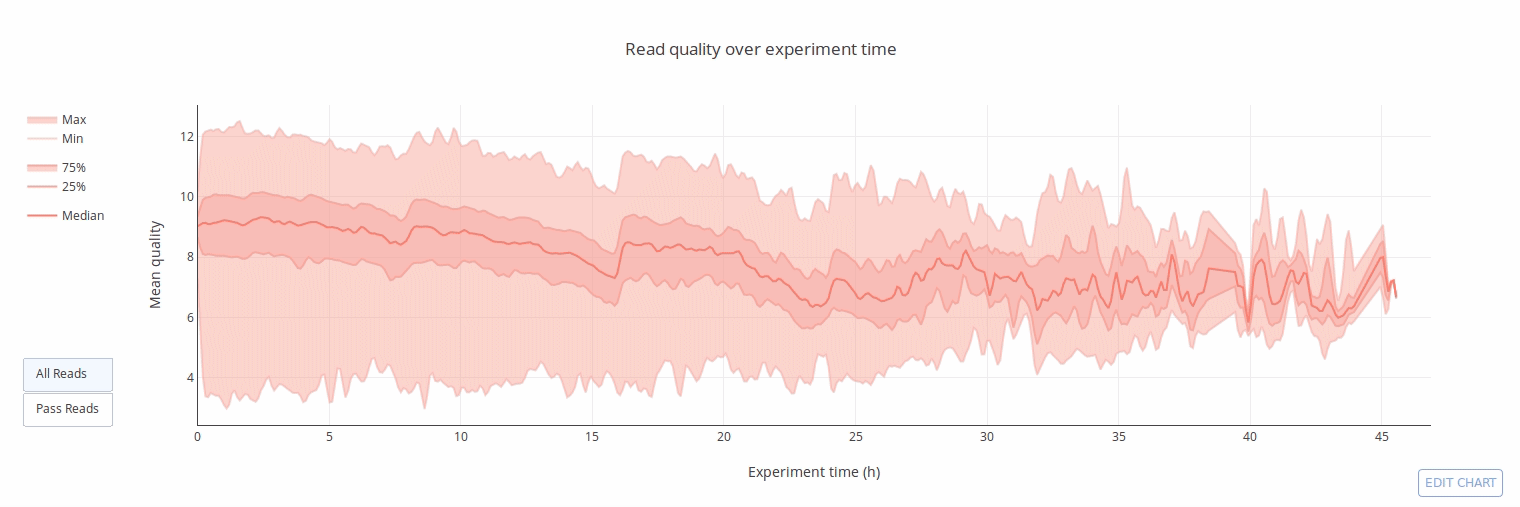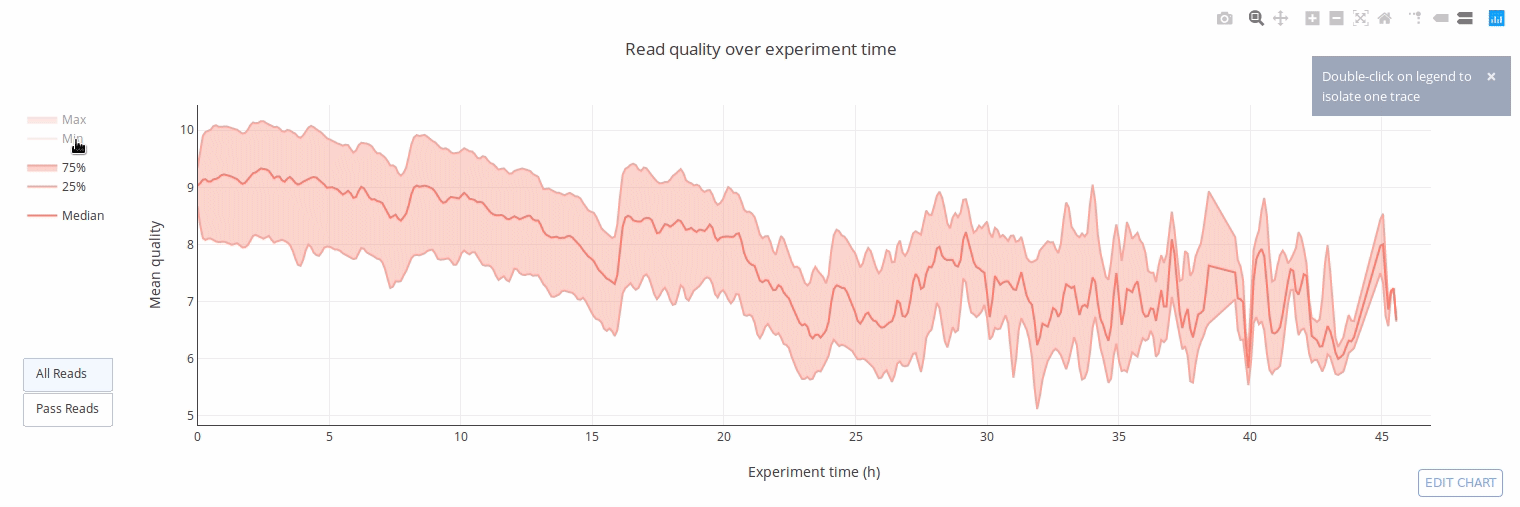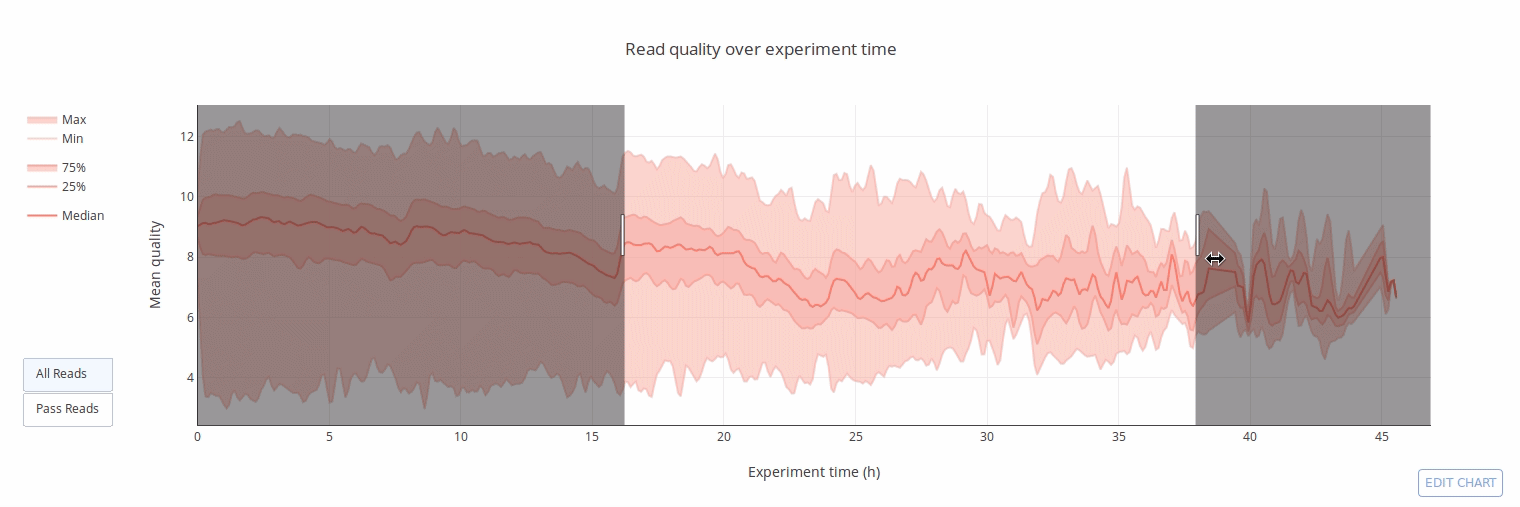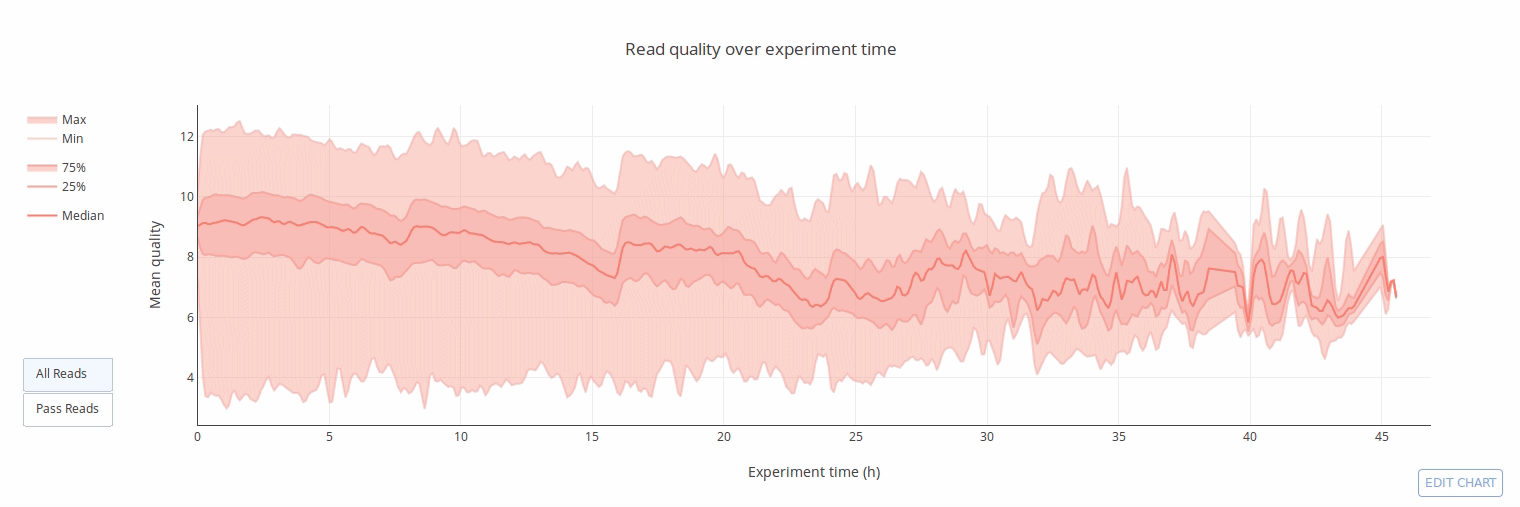
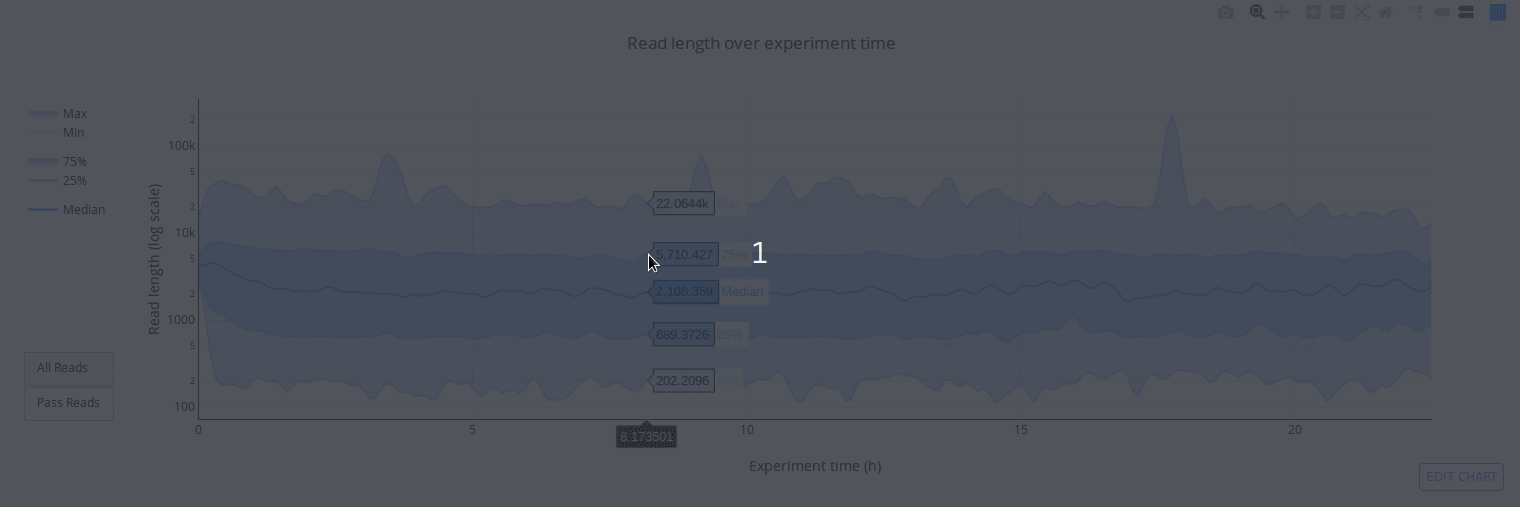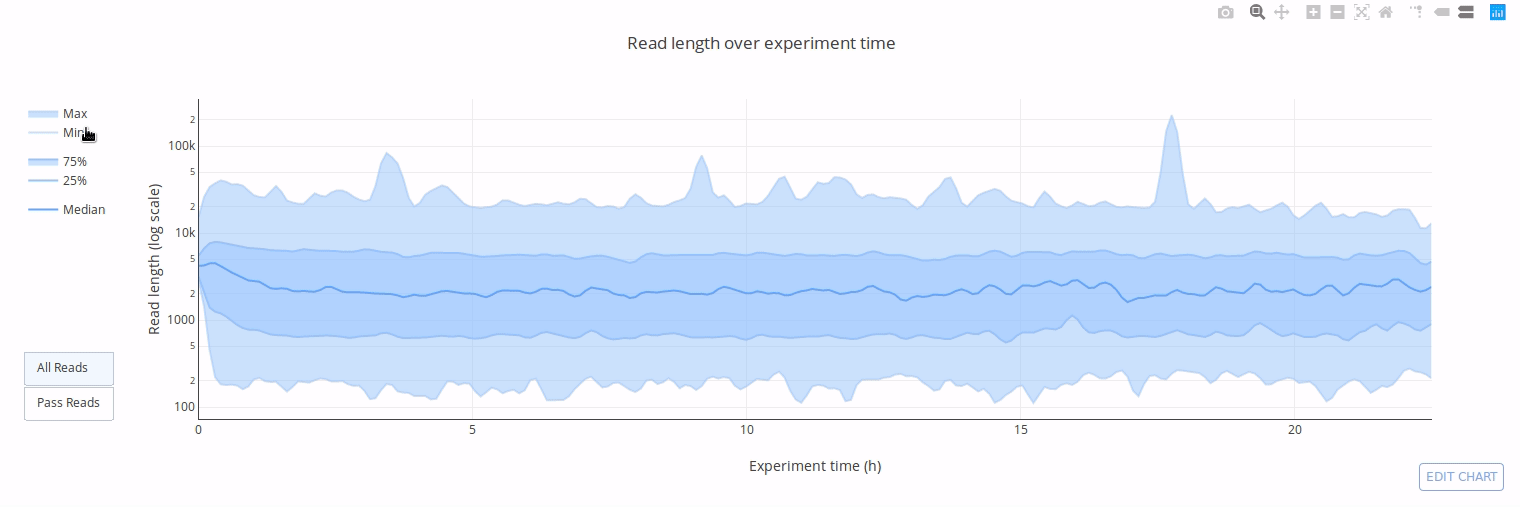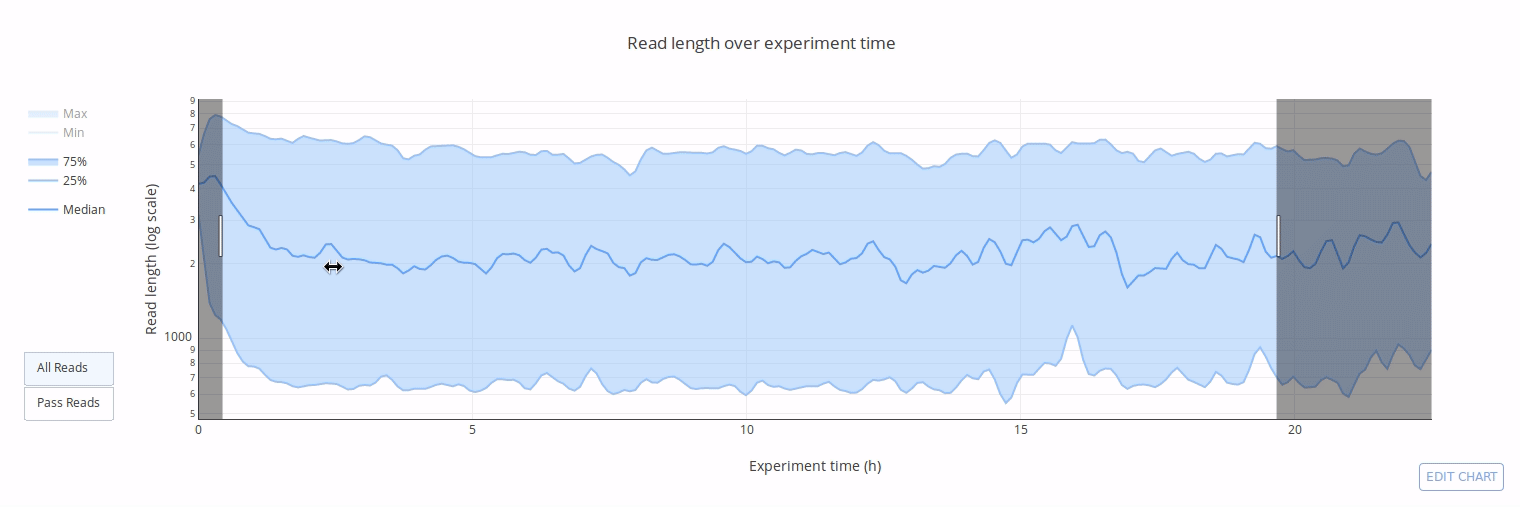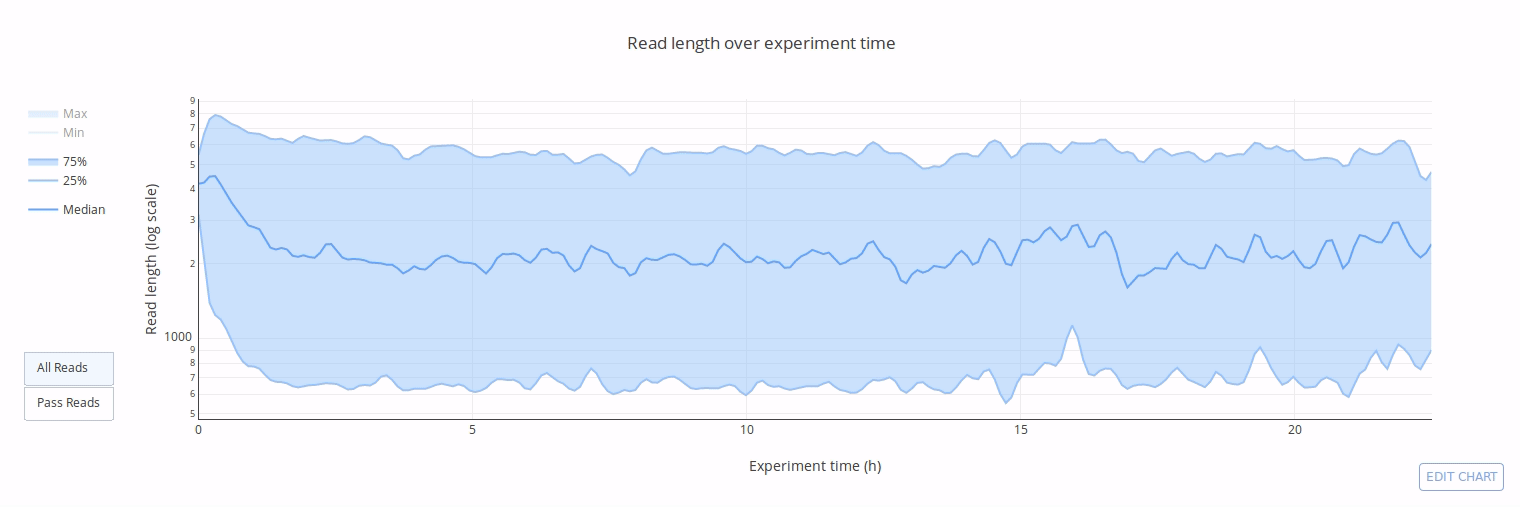
PycoQC computes metrics and generates interactive QC plots for Oxford Nanopore technologies sequencing data
PycoQC relies on the sequencing_summary.txt file generated by Albacore and Guppy, but if needed it can also generates a summary file from basecalled fast5 files. The package supports 1D and 1D2 runs generated with Minion, Gridion and Promethion devices and basecalled with Albacore 1.2.1+ or Guppy 2.1.3+. PycoQC is written in pure Python3. Python 2 is not supported.
Click on a picture to access an online interactive version editable with plotly chart studio
-
Adrien Leger - aleg {at} ebi.ac.uk
-
Tommaso Leonardi - tom {at} tleo.io