This repository contains the code to reproduce the results of the paper: "Guiding diffusion models for antibody sequence and structure co-design with developability properties".
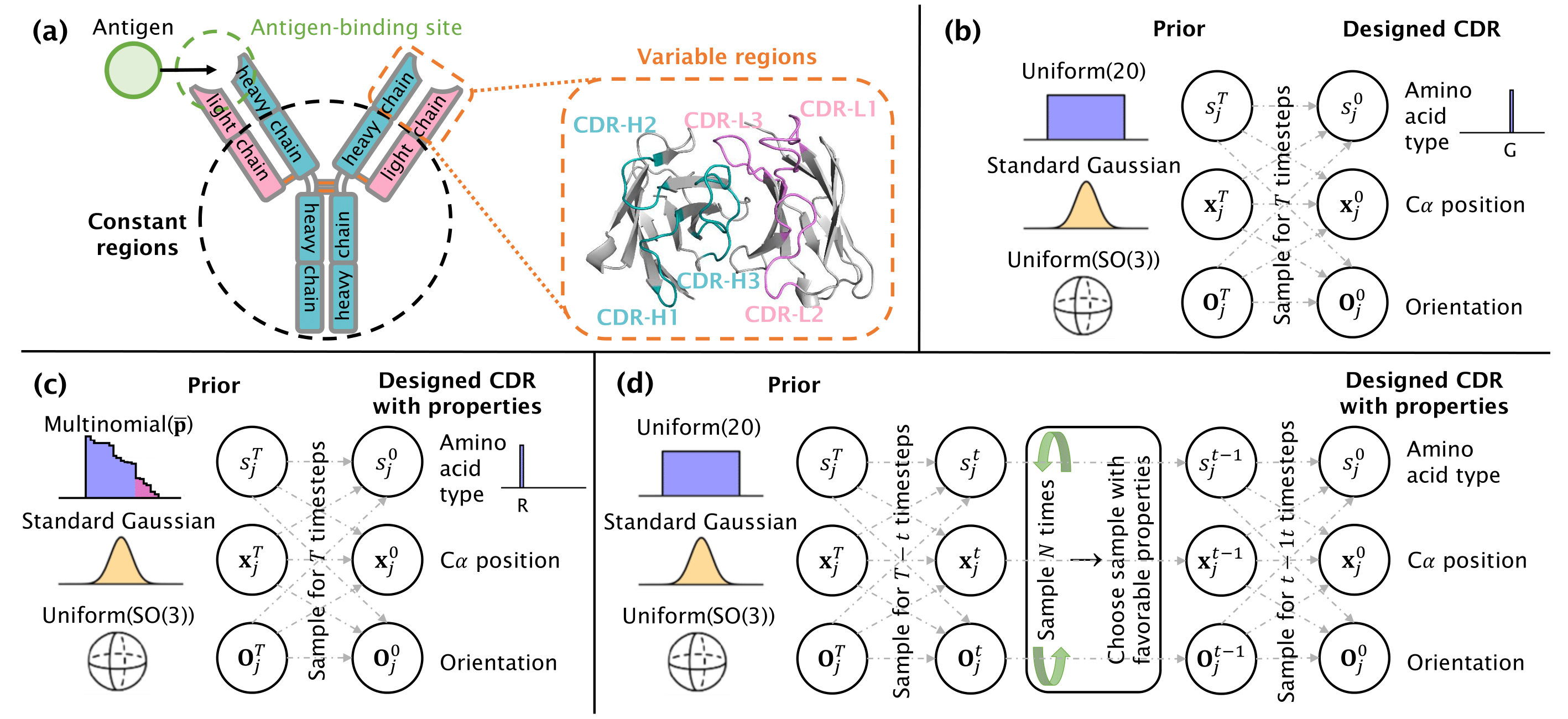 Figure: (a) Illustration of an antibody, featuring the two heavy (in blue) and two light (in pink) chains. The variable regions in the enlarged area encompass the antigen-binding site including the six CDR loops. (b-d) Visualization of the generative diffusion process, showcasing the prior distributions for each modality and the designed CDR, for the (b) property-unconditioned mode, (c) property-aware prior approach, and (d) sampling by property approach.
Figure: (a) Illustration of an antibody, featuring the two heavy (in blue) and two light (in pink) chains. The variable regions in the enlarged area encompass the antigen-binding site including the six CDR loops. (b-d) Visualization of the generative diffusion process, showcasing the prior distributions for each modality and the designed CDR, for the (b) property-unconditioned mode, (c) property-aware prior approach, and (d) sampling by property approach.
This repository builds upon (Luo et al. 2022) [DiffAb] and (Shan et al. 2022) [DDG Predictor]. To install the required packages, please refer to the documentation in the official repositories.
Protein structures in the SAbDab dataset for training and testing can be downloaded here. Extract all_structures.zip into the data folder. The data folder contains a snapshot of the dataset index (sabdab_summary_all.tsv).
[DiffAb] model weights can be downloaded from either [Hugging Face] or [Google Drive]. Copy the files into the trained_models folder. The model weights for [DDG Predictor] can be found in diffab/tools/ddg/data/model.pt.
The config files are in the configs/test folder. To design the six CDRs separately, use the codesign_single model and config on the scripts design_pdb.py (one sample) and design_testset.py (full test set, 19 samples). The lists of options are in the scripts diffab/tools/runner/design_for_pdb.py and diffab/tools/runner/design_for_testset.py, respectively.
# Sample 7DK2_AB_C
python design_pdb.py ./data/examples/7DK2_AB_C.pdb \
--config ./configs/test/codesign_single.yml
# Test set
for i in {0..18}; do
python design_testset.py $i --config ./configs/test/codesign_single.yml
doneFor hydropathy-aware prior with --prior_b as:
# Sample 7DK2_AB_C
python design_pdb.py ./data/examples/7DK2_AB_C.pdb \
--config ./configs/test/codesign_single.yml --prior_b 0.8
# Test set
for i in {0..18}; do
python design_testset.py $i \
--config ./configs/test/codesign_single.yml --prior_b 0.8
doneFor sampling by property (ddG, hydropathy, or both), use the following config files:
| Config file | Description |
|---|---|
codesign_single_ddg.yml |
Sequence-structure of one CDR, sampling by ddG. |
codesign_single_hydro.yml |
Sequence-structure of one CDR, sampling by hydropathy. |
codesign_single_ddg_and_hydro.yml |
Sequence-structure of one CDR, sampling by ddG and hydropathy. |
Here, the extra options are: --sample_step_mode ("min", "max", or "softmax"), --sample_step_num (int), and --sample_step_period (int). For example, for sampling by ddG use:
# Sample 7DK2_AB_C
python design_pdb.py ./data/examples/7DK2_AB_C.pdb \
--config ./configs/test/codesign_single_ddg.yml \
--sample_step_mode min --sample_step_num 20 --sample_step_period 1
# Test set
for i in {0..18}; do
python design_testset.py $i \
--config ./configs/test/codesign_single_ddg.yml \
--sample_step_mode min --sample_step_num 20 --sample_step_period 1
doneTo compute the evaluation metrics: AAR, RMSD, Hydropathy Score, and Predicted ddG (the option --no_energy prevents the computation of Rosetta ddG) for all samples use:
python eval.py --no_energy --root results/codesign_singleFor training details and more design options, such as "Fix-backbone Design", "Antibody Optimization", or "Antigen Only", please refer to the original implementation of [DiffAb].
If you find this repository useful in your research, please cite the following works.
@article{villegas2024guiding,
title={Guiding diffusion models for antibody sequence and structure co-design with developability properties},
author={Amelia Villegas-Morcillo and Jana M. Weber and Marcel J.T. Reinders},
journal={PRX Life},
volume={2},
number={3},
pages={033012},
year={2024}
}
@article{villegas2023guiding,
title={Guiding diffusion models for antibody sequence and structure co-design with developability properties},
author={Amelia Villegas-Morcillo and Jana M. Weber and Marcel J.T. Reinders},
journal={NeurIPS 2023 Generative AI and Biology Workshop},
year={2023},
url={https://openreview.net/forum?id=bPcgbKDCUQ}
}
@inproceedings{luo2022antigenspecific,
title={Antigen-Specific Antibody Design and Optimization with Diffusion-Based Generative Models for Protein Structures},
author={Shitong Luo and Yufeng Su and Xingang Peng and Sheng Wang and Jian Peng and Jianzhu Ma},
booktitle={Advances in Neural Information Processing Systems},
editor={Alice H. Oh and Alekh Agarwal and Danielle Belgrave and Kyunghyun Cho},
year={2022},
url={https://openreview.net/forum?id=jSorGn2Tjg}
}
@article{shan2022deep,
title={Deep learning guided optimization of human antibody against SARS-CoV-2 variants with broad neutralization},
author={Shan, Sisi and Luo, Shitong and Yang, Ziqing and Hong, Junxian and Su, Yufeng and Ding, Fan and Fu, Lili and Li, Chenyu and Chen, Peng and Ma, Jianzhu and others},
journal={Proceedings of the National Academy of Sciences},
volume={119},
number={11},
pages={e2122954119},
year={2022}
}