Xiuzhong Hu · Guangming Xiong · Zheng Zang · Peng Jia · Yuxuan Han · Junyi Ma
Beijing Institute of Technology
Our work has been accepted by IEEE Transactions on Intelligent Vehicles(TIV) and the paper link.
An early version for our work was: PC-NeRF: Parent-Child Neural Radiance Fields under Partial Sensor Data Loss in Autonomous Driving Environments.
The latest version on arxiv for our work is: PC-NeRF: Parent-Child Neural Radiance Fields Using Sparse LiDAR Frames in Autonomous.
@ARTICLE{10438873,
author={Hu, Xiuzhong and Xiong, Guangming and Zang, Zheng and Jia, Peng and Han, Yuxuan and Ma, Junyi},
journal={IEEE Transactions on Intelligent Vehicles},
title={PC-NeRF: Parent-Child Neural Radiance Fields Using Sparse LiDAR Frames in Autonomous Driving Environments},
year={2024},
volume={},
number={},
pages={1-14},
keywords={Laser radar;Three-dimensional displays;Robot sensing systems;Point cloud compression;Autonomous vehicles;Image reconstruction;Training;Neural Radiance Fields;3D Scene Reconstruction;Autonomous Driving},
doi={10.1109/TIV.2024.3366657}}[TOC]
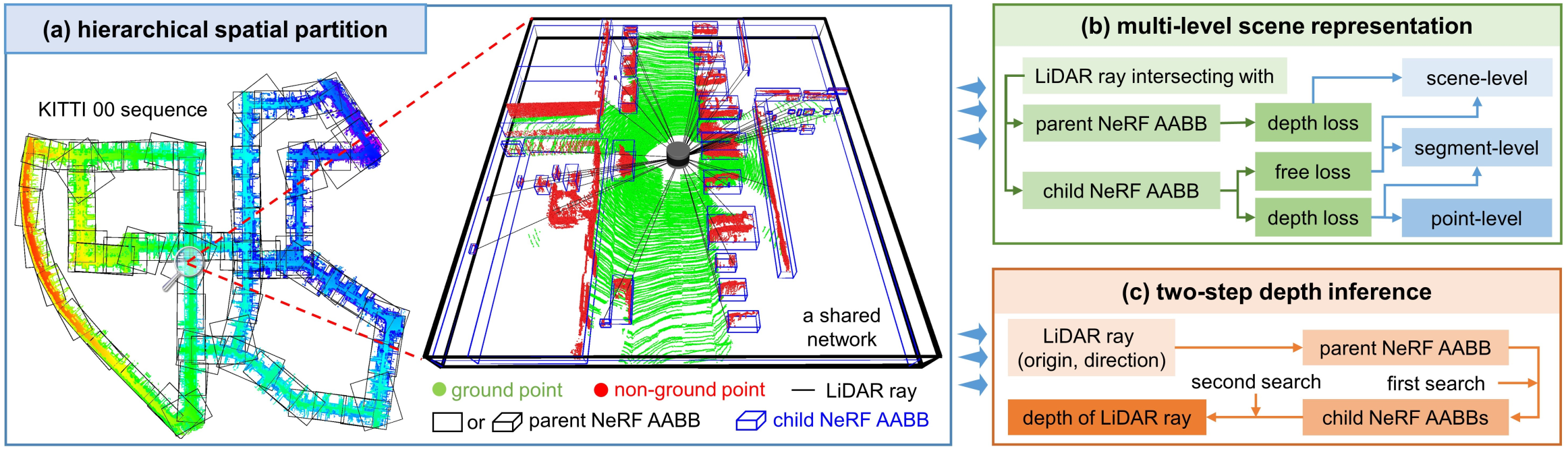
The framework for our work:
The performance of our work:
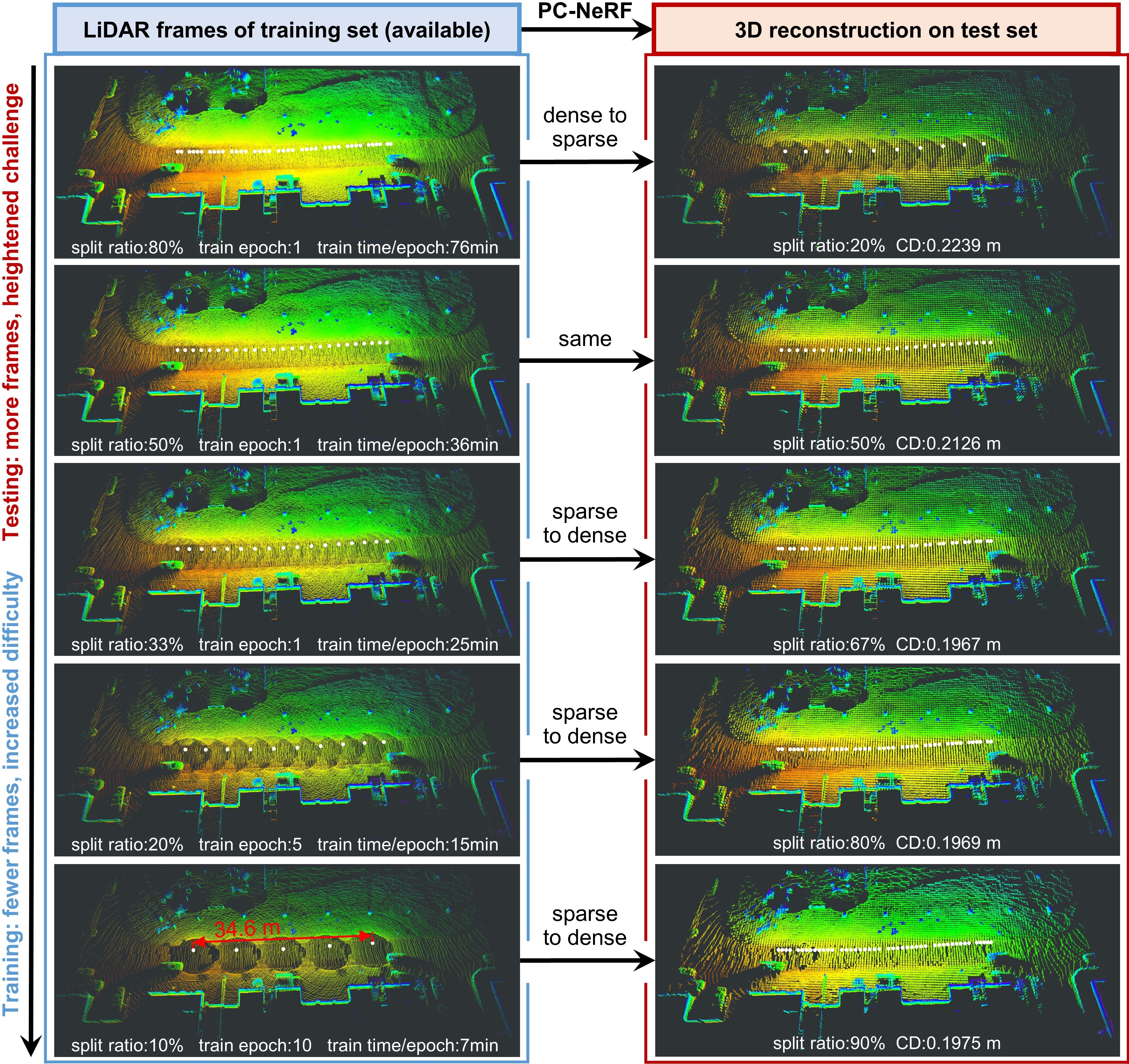
We use frame sparsity to measure the sparsity of LiDAR point cloud data in the temporal dimension. Frame sparsity represents the proportion of the test set (unavailable during training) when dividing the LiDAR dataset into training and test sets. Increased frame sparsity implies fewer LiDAR frames for training and more for model testing, posing heightened challenges across various tasks. For further details, please refer to Sec. IV-A of our paper.
In our work, we show the results at eight frame sparsities of {20 %, 25 %, 33 %, 50 %, 67 %, 75 %, 80 %, 90 %}. In the following, we demonstrate the process of dataset partitioning and data preprocessing with the frame sparsity of 20% as an example. When the frame sparsity is 20%, it means that one frame every five frames is selected as the test set and the rest is used as the training set.
The frame sparsity setting is reflected in the following three locations:
(1) in data pre-process of train dataset:
For example, in lines 52 to 56 of the data_preprocess/scripts/pointcloud_fusion.py file.
if((j+1-3)%5!=0): # test set: hold one form every 5 scans, so train set is the remain
count_train = count_train + 1
else:
continue For further details, please refer to the Data Pre-processing documentation.
(2) in train dataset load during training :
For example, in lines 646 to 660 of the nof/dataset/ipb2dmapping.py
file_path = os.path.join(self.root_dir, '{}.pcd'.format(j+1))
if((self.split == 'train') and (j+1-3-self.data_start)%5!=0): # frame sparsity = 20%
# if((self.split == 'train') and (j+1-self.data_start)%4!=0): # frame sparsity = 25%
# if((self.split == 'train') and (j+1-self.data_start)%3!=0): # frame sparsity = 33%
# if((self.split == 'train') and (j+1-self.data_start)%2!=0): # frame sparsity = 50%
# if((self.split == 'train') and (j+1-1-self.data_start)%3==0): # frame sparsity = 67%
# if((self.split == 'train') and (j+1-1-self.data_start)%4==0): # frame sparsity = 75%
# if((self.split == 'train') and (j+1-3-self.data_start)%5==0): # frame sparsity = 80%
# if((self.split == 'train') and (j+1-5-self.data_start)%10==0): # frame sparsity = 90%
print("train file_path: ",file_path)
elif (self.split == 'val' and (j+1-3)%5==0):
# elif (self.split == 'val' and 0 ): # Note: More stringent conditions can be set to make the validation set empty
print("val file_path: ",file_path)
else:
continue (3) in test dataset load during testing :
For example, in lines 1054 to 1062 of the eval_kitti_render.py
for j in range(hparams.data_start, hparams.data_end):
if((j+1-3-hparams.data_start)%5==0): # frame sparsity = 20%
# if((j+1-hparams.data_start)%4==0): # frame sparsity = 25%
# if((j+1-hparams.data_start)%3==0): # frame sparsity = 33%
# if((j+1-hparams.data_start)%2==0): # frame sparsity = 50%
# if((j+1-1-hparams.data_start)%3!=0): # frame sparsity = 67%
# if((j+1-1-hparams.data_start)%4!=0): # frame sparsity = 75%
# if((j+1-3-hparams.data_start)%5!=0): # frame sparsity = 80%
# if((j+1-5-hparams.data_start)%10!=0): # frame sparsity = 90%γ is a constant designed to represent the smooth transition interval on a LiDAR ray between the child NeRF free loss and the child NeRF depth loss.
γ is a parameter that cannot currently be adjusted by bash scripts, but can be set in bash scripts via parameter passing.
The expand_threshold in ./nof/render.py is used to adjust γ.
# (line 91~99)
for i, row in enumerate(z_vals):
expand_threshold = 2 #
interval = near_far_child[i]
mask_in_child_nerf[i] = (interval[0]-expand_threshold <= row) & (row <= interval[1]+expand_threshold)
while (abs(torch.sum(mask_in_child_nerf[i]))==0): #
expand_threshold = expand_threshold + 0.01
mask_in_child_nerf[i] = (interval[0]-expand_threshold <= row) & (row <= interval[1]+expand_threshold)
weights_child = weights * mask_in_child_nerf.float() #
z_vals_child= z_vals * mask_in_child_nerf.float() # In the following experiments the frame sparsity is 20%, and the experiments for other frame sparsities are similar.
In data/preprocessing/,logs/kitti00/1151_1200_view/save_npy/split_child_nerf2_3/,logs/maicity00/maicity_00_1/save_npy/split_child_nerf2_3/,data_preprocess/kitti_pre_processed/sequence00/, part of the file exceeds the github recommended size for a single file, and the download link.
.
├── data
│ └── preprocessing
│ ├── kitti00
│ └── maicity00
├── data_preprocess
│ └── kitti_pre_processed
│ └── sequence00
└── logs
├── kitti00
│ └── 1151_1200_view
│ └── save_npy
│ └── split_child_nerf2_3
└── maicity00
└── maicity_00_1
└── save_npy
└── split_child_nerf2_3conda create -n pcnerf python=3.9.13
conda activate pcnerf
conda install -c conda-forge pybind11
# RTX3090
conda install pytorch==1.12.1 torchvision==0.13.1 torchaudio==0.12.1 cudatoolkit=11.3 -c pytorch
pip install pytorch-lightning tensorboardX
pip install matplotlib scipy open3d
pip install evo --upgrade --no-binary evo# train
# epoch = 1
bash ./shells/pretraining/KITTI00_originalnerf_train.bash
# eval
# depth_inference_method = 1: use one-step depth inference
# depth_inference_method = 2: use two-step depth inference
# test_data_create = 0: use the test data created by ours
# test_data_create = 1: recreate the test data
bash ./shells/pretraining/KITTI00_originalnerf_eval.bash# train
# epoch = 1
bash ./shells/pretraining/KITTI00_pcnerf_train.bash
# eval
# depth_inference_method = 1: use one-step depth inference
# depth_inference_method = 2: use two-step depth inference
# test_data_create = 0: use the test data created by ours
# test_data_create = 1: recreate the test data
bash ./shells/pretraining/KITTI00_pcnerf_eval.bash# inference_type: one-step: print the metric of one-step depth inference
# inference_type: two-step: print the metric of two-step depth inference
# version_id: version_0 (OriginalNeRF) or version_1 (PC-NeRF)
cd ./logs/kitti00/1151_1200_view/render_result/
python print_metrics.py# train
# epoch = 1
bash ./shells/pretraining/MaiCity00_originalnerf_train.bash
# eval
# depth_inference_method = 1: use one-step depth inference
# depth_inference_method = 2: use two-step depth inference
# test_data_create = 0: use the test data created by ours
# test_data_create = 1: recreate the test data
bash ./shells/pretraining/MaiCity00_originalnerf_eval.bash# train
# epoch = 1
bash ./shells/pretraining/MaiCity00_pcnerf_train.bash
# eval
# depth_inference_method = 1: use one-step depth inference
# depth_inference_method = 2: use two-step depth inference
# test_data_create = 0: use the test data created by ours
# test_data_create = 1: recreate the test data
bash ./shells/pretraining/MaiCity00_pcnerf_eval.bash# inference_type: one-step: print the metric of one-step depth inference
# inference_type: two-step: print the metric of two-step depth inference
# version_id: version_0 (OriginalNeRF) or version_1 (PC-NeRF)
cd ./logs/maicity00/maicity_00_1/render_result/
python print_metrics.pyThe following codes have helped us a lot in our work:nerf-pytorch, ir-mcl , shine_mapping