This tool is no longer under development, please use brainglobe-napari-io instead
Visualise cellfinder results with napari
You can install napari-cellfinder via pip:
pip install napari-cellfinder
- Open napari (however you normally do it, but typically just type
napariinto your terminal) - Load your raw data (drag and drop the data directories into napari, one at a time)
- Drag and drop your cellfinder output directory into napari.
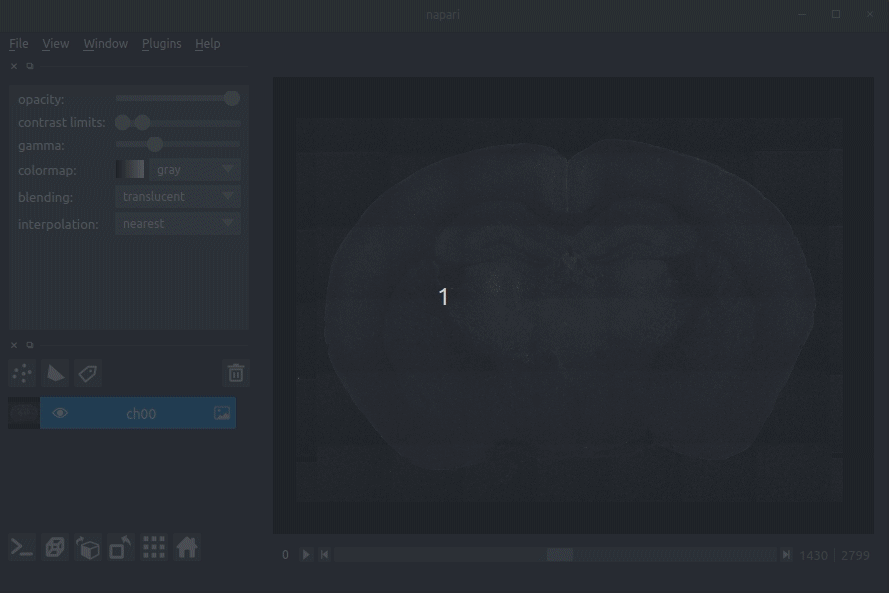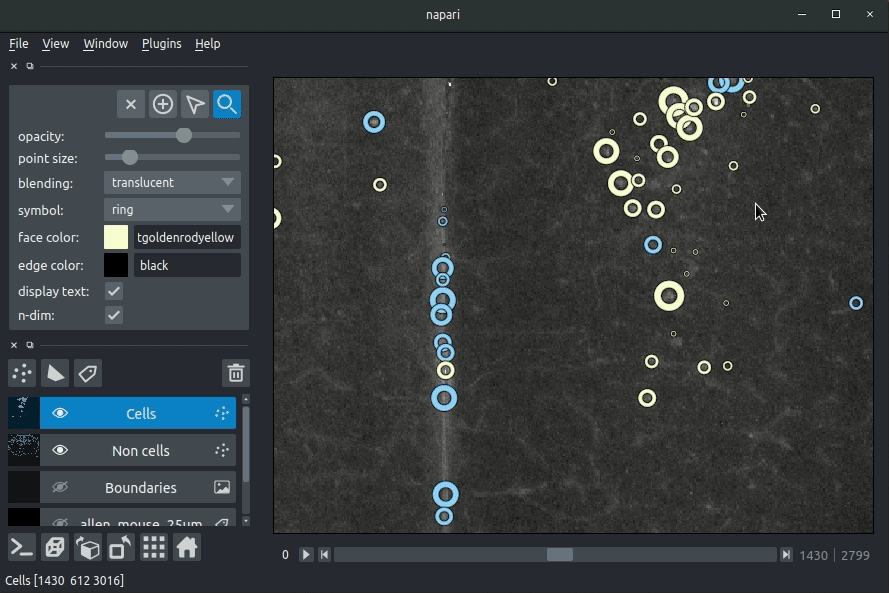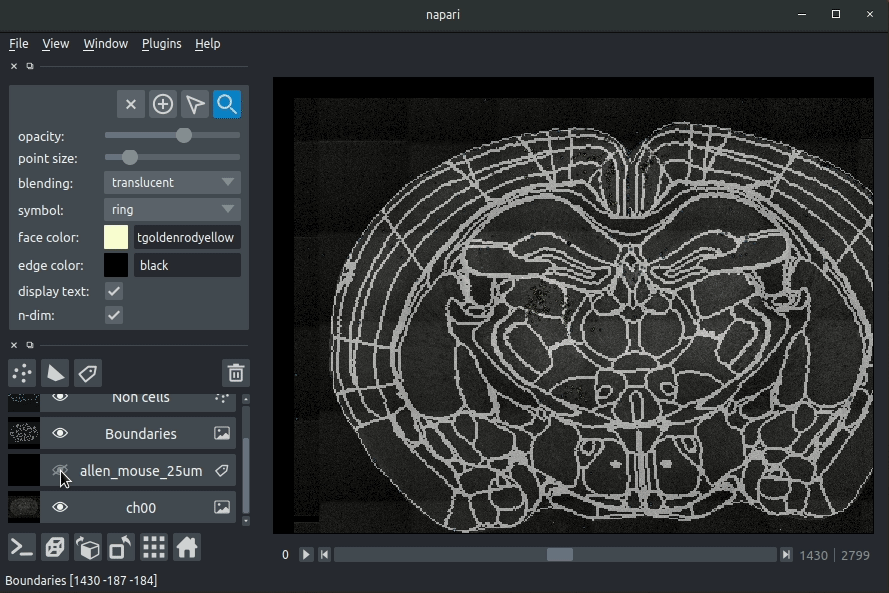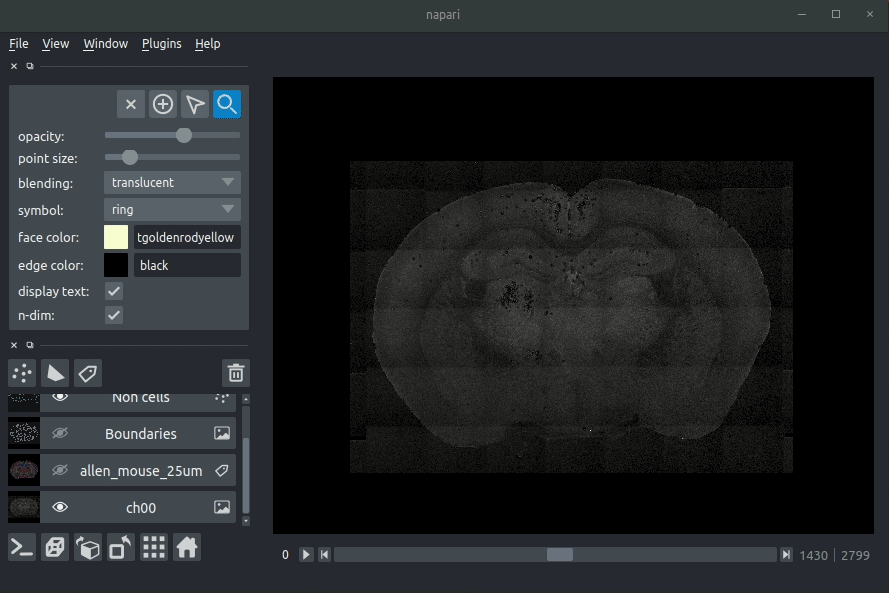
The plugin will then load your detected cells (in yellow) and the rejected cell candidates (in blue). If you carried out registration, then these results will be overlaid (similarly to the napari-brainreg plugin, but transformed to the coordinate space of your raw data).
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
Distributed under the terms of the MIT license, "napari-cellfinder" is free and open source software
If you encounter any problems, please file an issue along with a detailed description.