This pipeline is designed to take reads from Illumina or Nanopore data and generate any of the following:
- Alignments
- Variant calls
- Consensus sequences from variant calls
- TSVs to easily import variant calls into R for analysis
- Multiple sequence alignments
Important
When a line in a code block starts with a $, it means that that line should be entered into the terminal prompt. Do not include the $ when typing/copying the command into the prompt.
I will also embed anything that should be filled in in <these brackets>. Don't include the brackets when you fill in your own information.
I HIGHLY recommend learning some basic terminal skills before attempting this tutorial. I will show step by step instructions for how to run the pipeline, but will not include a tutorial for basic terminal navigation skills. This is just one guide, but there are many guides out there you can use as a reference.
Note
This guide is designed for use with Unix systems (Mac, Linux). Windows users may need to use Windows Subsystem for Linux (WSL) for some commands to work.
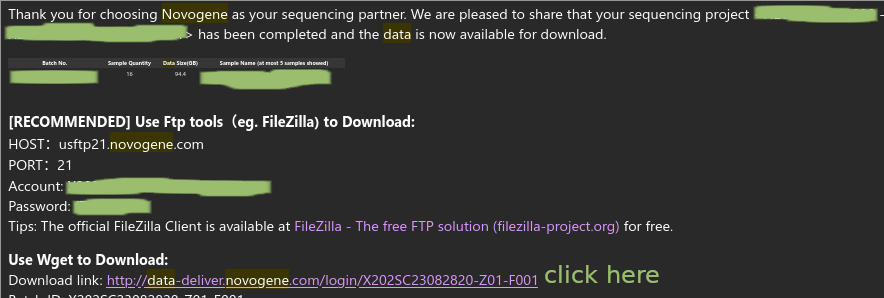
When your data is ready, you should get an email that looks like this. Click the link highlighted in yellow:
It should ask you for your email and password (you used these to make an account to put in the order).
Go to the data release for your project and click Batch download. A panel will open up on the side with a wget command. In the terminal, connect to Longleaf and go to the directory in which you plan to store your data. Send the wget command as a job to SLURM, Longleaf's job handling application. This will take a while.
# connect to Longleaf
$ ssh <onyen>@longleaf.unc.edu
# go to the directory where you want to store the data
$ cd /proj/sekellab/projects/<PROJECT-NAME>/
# submit the wget command as a job to SLURM
$ sbatch -p general -N 1 -n 1 --mem=5g -t 05-00:00:00 --wrap="<WGET-COMMAND-FROM-NOVOGENE>"
You should be in the directory with your Novogene data, and your directory should now look something like this (with sample being a placeholder for a sample name):
$ tree
.
└── usftp21.novogene.com/
├── 01.RawData/
│ └── sample/
│ ├── MD5.txt
│ ├── sample_CKDN230029067-1A_HCTF3DSX7_L3_1.fq.gz
│ ├── sample_CKDN230029067-1A_HCTF3DSX7_L3_2.fq.gz
│ ├── sample_CKDN230029067-1A_HGKT2DSX7_L4_1.fq.gz
│ └── sample_CKDN230029067-1A_HGKT2DSX7_L4_2.fq.gz
├── 02.Report_X202SC23082820-Z01-F001.zip
├── checkSize.xls
├── MD5.txt
├── MD5.zip
└── Readme.html
Note
The number of files and the suffix for each of the sample files (e.g. CKDN230029067-1A_HCTF3DSX7) will vary.
Tip
You can use the command tree to display a directory structure like the one above.
You should check the checksum of all of your files to make sure the downloads were complete.
$ cd usftp21.novogene.com
$ md5sum -c MD5.txt
If it works, it will say something like:
01.RawData/sample/sample_CKDN230029067-1A_HCTF3DSX7_L3_1.fq.gz: OK
# ... and so on for every file in all of the sample directories
Note
If the md5sum check fails, your data is corrupted and you need to redownload it.
Because the file size to transfer is large, you will need to use GLOBUS to transfer the data from the Nanopore computer to Longleaf.
First, compress the data to make it easier to transfer. Because /var isn't a subdirectory of your home directory, you will need to perform this operation as the root user (or "super user", sudo = Super User Do. This is like running a program with admin privileges, in Windows). Since you are performing this command as root, it will prompt you for the computer's password.
$ cd /var/lib/minknow/data/<PROJECT-NAME>/<FLOWCELL-ID>
$ sudo tar -czvf <PROJECT-NAME>.tar.gz fastq_pass/*
You should now have a file called <PROJECT-NAME>.tar.gz (where PROJECT-NAME is a placeholder for whatever you named your project in MinKNOW).
GLOBUS can't recognize any directories above your home directory, so let's move the compressed file somewhere else:
$ sudo mv <PROJECT-NAME>.tar.gz ~
Now <PROJECT-NAME>.tar.gz is in our home directory (~).
Start up Globus Connect Personal on the Nanopore computer:
$ ~/globusconnectpersonal & disown
A window should pop up. Click "Connect" and make sure not to close the window.
In a web browser, go to https://www.globus.org and click "Log in" at the top right. Use the organizational login, and search for University of North Carolina - Chapel Hill in the drop down. There will be a few screens to go through and then you'll be prompted for your ONYEN. See https://help.rc.unc.edu/getting-started-with-globus-connect for more detail and screenshots.
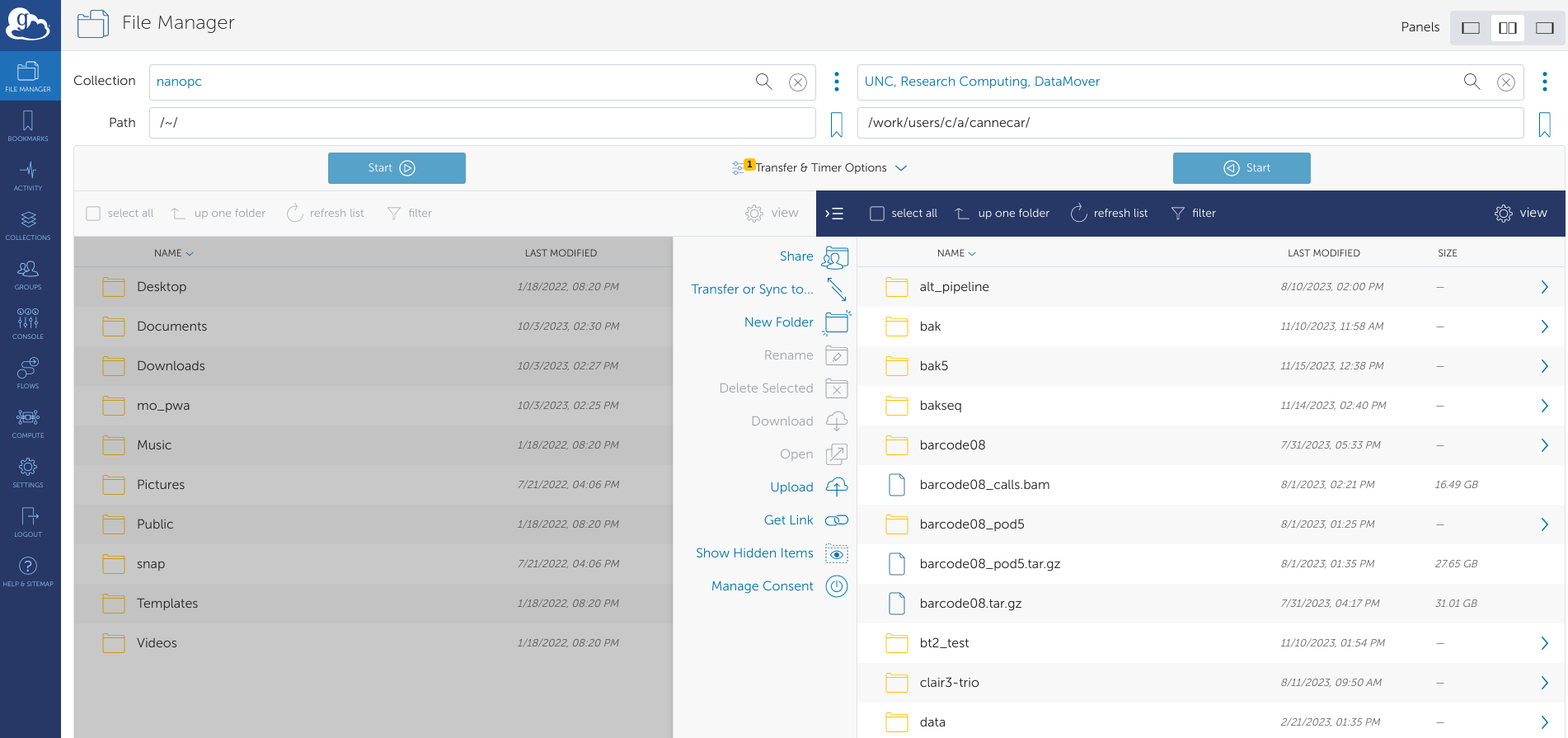
Once you're logged in, click the File Manager tab on the left side. You'll see a split screen view of two different data collections:
In the left collection search box, type "nanopc" and click the result that pops up. This is the Nanopore computer. In the right box, search "UNC, Research Computing, DataMover" and click the result that pops up. This is Longleaf.
Note
If the icon for nanopc is red, indicating that the connection is offline, start Globus Connect Personal again by typing ~/globusconnectpersonal in the terminal.
You will automatically be in the home directory for both collections. In the UNC DataMover collection, change the path to /work/users/<o>/<n>/<onyen>/.
Find and click your <PROJECT-NAME>.tar.gz file in the left collection (it should be highlighted in blue), then click the blue Start button on the left. A green popup should appear saying the transfer request has been submitted.
Now click the Activity panel on the left side menu. You should find an entry that says "nanopc to UNC, Research Computing, DataMover". Click it,and it will take you to a page showing the progress of your transfer.
When the condition says "SUCCEEDED" your file transfer has finished.
We are now ready to clone the pipeline.
If you haven't already, go to the terminal and connect to Longleaf via SSH:
$ ssh <onyen>@longleaf.unc.edu
Go to your work directory on Longleaf at /work/users/<o>/<n>/<onyen>/ (where 'o' and 'n' are the first and second letters of your ONYEN, respectively).
$ cd /work/users/<o>/<n>/<onyen>/
Clone the pipeline from here (click the green button that says Code, then copy the link in the drop down), then go to the directory of the repository.
$ git clone https://github.com/caturcotte/seq_pipeline.git
$ cd seq_pipeline/
The directory tree of the pipeline should look like this:
$ tree
.
├── config.yaml
├── miniforge_installer.sh
├── README.md
├── sample_sheet.csv
├── slurm
│ └── config.yaml
├── slurm_sub.sh
├── Snakefile
└── workflow
├── alignment.smk
├── calling.smk
├── envs
│ ├── bcftools.yaml
│ ├── bowtie2.yaml
│ ├── bwa.yaml
│ ├── concat_vcfs.yaml
│ ├── environment.yaml
│ ├── freebayes.yaml
│ ├── minimap2.yaml
│ ├── repeatmasker.yaml
│ └── samtools.yaml
├── functions.smk
├── misc.smk
├── qc.smk
└── vcf_filtering.smk
Now that the pipeline has been copied to Longleaf, we can configure it.
Note
There are two files called config.yaml - one in seq_pipeline/ and one in seq_pipeline/slurm/. Here I am referring to the one in seq_pipeline/.
Open config.yaml in a text editor:
$ nano config.yaml
You will see that there are numerous options for configuring the pipeline for your needs. There are comments in the file explaining the purpose of all of the options. Some of the more notable ones:
ref_nameandref_file: The name and location of the reference genome FASTA, respectively. The default is the dm6 release from UCSC, which is already stored on Longleaf. The pipeline will symlink (make a shortcut) to your ref_file atdata/resources/ref_name.fa. Note that the original files are not altered in the pipeline, so there isn't a need to make a copy.data_locations: These are shorthands for specific locations where your data is located. See the section on pointing the pipeline to your read files for more detail.
In a new terminal, move to a directory you can easily access via your file browser. In this directory, open up an SFTP connection to Longleaf and copy sample_sheet.csv to your local computer.
$ cd ~/Downloads
$ sftp <onyen>@longleaf.unc.edu
$ cd /work/users/<o>/<n>/<onyen>/seq_pipeline/
$ get sample_sheet.csv
In this case, the file was downloaded to the Downloads folder.
Tip
If you open your SFTP connection in the wrong location, you can use lls (local list directory) and lcd (local change directory) to navigate to a better location without exiting the connection.
Edit sample_sheet.csv in Excel. Each of your samples will have its own row in the sheet.
Note
The adapter fields (Novogene and Nanopore) and barcode field (for Novogene only) can be left blank. If left blank, adapters/barcodes will not be trimmed for that sample. Often reads come demultiplexed, meaning reads containing adapter/barcodes have already been trimmed because the barcode sequences were used to sort the files into the folders for their respective samples.
For Novogene samples the sample names must match the original names you gave the samples when you sent them to Novogene.
Tip
You can check your sample names with ls /path/to/reads/usftp21.novogene.com/01.RawData/ - all of the subdirectories in this directory will correspond to the names you gave your samples originally.
For Nanopore samples the sample name can be whatever you want so long as it does not include spaces, but the barcode column should contain the number of the barcode used for that sample.
Save sample_sheet.csv as a csv file under the same name and directory as before (rewrite the original file). Then re-upload the file to Longleaf:
$ put sample_sheet.csv
You can now exit the SFTP connection (type exit) and go back to your original SSH connection (if you saved the other terminal window, otherwise connect to Longleaf and change directory to the seq_pipeline folder again).
Note
Multiple locations can be specified in data_locations. Each location should be on a new line in the file.
The way to point the pipeline to your files differs slightly between Novogene and Nanopore data.
Here I show examples for both. In both cases we will assume the data_locations entry in config.yaml looks like this, with an entry called example_location:
data_locations:
example_location: /absolute/path/to/files/
If, in sample_sheet.csv, the location of sample A is set to example_location:
| sample | group | platform | source | read_type | location |
|---|---|---|---|---|---|
| A | group1 | illumina | novogene | paired | example_location |
The pipeline will look for these files for A:
/absolute/path/to/files/usftp21.novogene.com/01.RawData/A/A*_1.fq.gz (read 1) and /absolute/path/to/files/usftp21.novogene.com/01.RawData/A/A*_2.fq.gz (read 2).
Note
* is a wildcard meaning "one or more occurances of any character." For example, A*_1.fq.gz will match all read 1 files for sample A.
If, in sample_sheet.csv, the location of sample A is set to example_location:
| sample | group | platform | source | read_type | location | barcode |
|---|---|---|---|---|---|---|
| A | group1 | nanopore | nanopore | single | example_location | 1 |
The pipeline will look for the files for A in /absolute/path/to/files/fastq_pass/barcode01/*.fq.gz.
Open slurm_sub.sh:
$ nano slurm_sub.sh
Edit the mail-user line, replacing YOUR-EMAIL-HERE with your UNC email.
Save and exit the file with Ctrl+O and Ctrl+X.
Do the same for slurm/config.yaml.
We're ready to run the pipeline! Make sure you're in seq_pipeline, then use the following command to run the pipeline:
$ sbatch slurm_sub.sh
The output should say Submitted batch job JOB-NUMBER.
The pipeline is now running. While it's running, you can monitor the status of your jobs.
You can do the following to monitor the status of your jobs:
$ squeue -u <onyen>
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
The pipeline will submit jobs to SLURM automatically through your account, so you will likely see several jobs here. ST indicates the status of the job, where PD means pending and R means that the job is currently running. TIME indicates the time that the job has been running for.
If you want to continually monitor the status of your jobs:
$ squeue -u <onyen> -i 60
This will update with the status of your jobs every minute (press Ctrl+C to cancel).
To continually follow the output of your jobs, use:
$ tail -F slurm-<JOB-NUMBER>.out
This will continually report the status of all of the jobs (press Ctrl+C to cancel).
If you see an error, such as:
[Mon Nov 27 10:41:49 2023]
Error in rule sort_bams:
jobid: 15
input: data/alignments/ndj_01_CKDN230029052-1A_HGL2TDSX7_L1.bam
output: data/alignments/ndj_01_CKDN230029052-1A_HGL2TDSX7_L1_sort.bam
shell:
samtools sort data/alignments/ndj_01_CKDN230029052-1A_HGL2TDSX7_L1.bam -l 1 -o data/alignments/ndj_01_CKDN230029052-1A_HGL2TDSX7_L1_sort.bam --threads 4
(one of the commands exited with non-zero exit code; note that snakemake uses bash strict mode!)
cluster_jobid: Submitted batch job 23700119
Error executing rule sort_bams on cluster (jobid: 15, external: Submitted batch job 23700119, jobscript: /work/users/c/a/cannecar/seq_pipeline/.snakemake/tmp.x1vvuyv2/snakejob.sort_bams.15.sh). For error details see the cluster log and the log files of the involved rule(s).
Then there was a problem with a job. You can view the output of one of the jobs that failed to see more specific details (see below).
Tip
If one of the jobs gave you an error. you can use the error message from the main output log to quickly find the corresponding log file.
Let's say the error message from the main output logis "Error executing rule format_bcf on cluster (jobid: 26, external: Submitted batch job 23966205".
In seq_pipeline/ you can find the corresponding log file with find logs/ -name '23966205' (which will print the name of the file to the terminal output).
In the logs/ folder, there will be folders for each job that has been run. For example:
$ cd /work/users/c/a/cannecar/seq_pipeline/logs/
$ ls
align_bwa bwa_idx fix_mate_pairs mask_repeats separate_into_samples
bcftools_call bwa_mem format_bcf merge_bams sort_bams
bcftools_index faidx_ref freebayes merge_tsvs vcf_stats
bcftools_mpileup_single fastqc_paired index_bams mosdepth
bcftools_norm filter_low_quality mark_duplicates multiqc
If we go into one of these folders, there will be a file for each sample. named in the form SLURM-JOB-ID-JOB-NAME-sample=SAMPLE.out:
$ cd merge_bams
$ ls
'merge_bams-sample=ndj_01.out' 'merge_bams-sample=ndj_07.out' 'merge_bams-sample=ndj_13.out'
'merge_bams-sample=ndj_02.out' 'merge_bams-sample=ndj_08.out' 'merge_bams-sample=ndj_14.out'
'merge_bams-sample=ndj_03.out' 'merge_bams-sample=ndj_09.out' 'merge_bams-sample=ndj_15.out'
'merge_bams-sample=ndj_04.out' 'merge_bams-sample=ndj_10.out' 'merge_bams-sample=ndj_16.out'
'merge_bams-sample=ndj_05.out' 'merge_bams-sample=ndj_11.out'
'merge_bams-sample=ndj_06.out' 'merge_bams-sample=ndj_12.out'
You can view the output of one of these files using cat FILE-NAME (which prints the contents to stdout, the terminal output) or less FILE-NAME (which opens the contents in a separate window - press q to return to the terminal prompt. If you're wondering why it's called less, it's because less is the improved successor of a more suitably named program called more, which does the same thing... so "less is more").
If the job completed successfully, the end of the file will probably say something like
[Tue Nov 28 11:26:26 2023]
Finished job 0.
1 of 1 steps (100%) done
If not, the job is either still running or there will be some error you can see in the log file.
All of the output files should be in the data/ directory:
| output type | location | recommended tool(s) to view |
|---|---|---|
| alignments | data/alignments/ | IGV |
| calls | data/calls/ | bcftools, less, IGV |
| consensus | data/consensus/ | Snapgene, IGV |
| multiple sequence alignments | data/msa | Snapgene, IGV |
| tsvs | data/tsvs | R |
You can use IGV on Longleaf directly so long as you use the -X flag when establishing your SSH connection:
ssh -X <onyen>@longleaf.unc.edu
Note
To use the -X flag on Mac you'll need to download XQuartz.
The pipeline will automatically run several QC metrics for your samples and compile them into one report. To view the report, it's easiest to download the html content first to your home computer.
Exit your SSH connection and connect to Longleaf via SFTP. Then, navigate to the data/qc folder within seq_pipeline and download the multiqc files:
$ sftp <onyen>@longleaf.unc.edu
$ cd /work/users/<o>/<n>/<onyen>/seq_pipeline/data/qc/
$ get -r multiqc*
# files will download...
$ exit
Now, open multiqc.html in Firefox:
$ firefox multiqc.html
There are various metrics in this report that you can look at. A good one to start with is the FASTQC report, which tells you the quality of your reads. You can also see the average read depth for each of your samples, and transition to transversion ratio to determine the quality of the calls. For Drosophila WGS, the Ts/Tv ratio should be around 2 [1]. Too low indicates a high false positive rate of calling, and too high indicates a biased callset (see this post)
See here for more information on variant calling metrics.
Alignment and BAM processing
First, all of the reads or read pairs for a given sample will be individually aligned to whatever reference genome is selected in config.yaml. The alignments will then be sorted and merged into a single BAM file, with lane information being preserved in the read group tags of the reads. Next, the BAM files will be processed to remove duplicates. This is only necessary for Illumina sequencing and sequencing of PCR-amplified DNA, but it will not be detrimental for analysis of Nanopore reads.
flowchart TD;
A[sample 1]--- a{{sequencing}}-->B[sample 1 lane 1 reads] & C[sample 1 lane 2 reads] & D[sample 1 lane 3 reads]
B --- b{{alignment}} -->E[sample 1 lane 1 BAM]
C --- c{{alignment}} -->F[sample 1 lane 2 BAM]
D --- d{{alignment}} -->G[sample 1 lane 3 BAM]
E & F & G --> q{{sort and merge}} --> H[sample 1 BAM]
H --- f{{remove duplicates}} --> I[sample 1 BAM, duplicates removed]
Next, the polished BAM files (with duplicates removed) will be pooled into the groups specified in the sample sheet. Variants will be called and sorted into variant call format files.
flowchart TD
H[sample 1 BAM] & I[sample 2 BAM] & J[sample 3 BAM]---z{group 1}---e{{variant calling}} -->K[VCF]
K --- a{{filtering}} --> L[filtered VCF] ---r{{separate by sample}} -->M[sample 1 VCF] & N[sample 2 VCF] & O[sample 3 VCF]
P[sample 4 BAM] & Q[sample 5 BAM]---g{group 2}-->k[the same process, separately]
Using these VCFs, you can make consensus sequences for your samples by applying the variants in the sample to the reference genome. The resulting FASTA files can be viewed in Snapgene, etc. for downstream analysis.
flowchart TD
A[sample 1 VCF] & B[reference FASTA] ---q{{make consensus sequence}}-->C[sample 1 reference FASTA]
Consensus sequences can be aligned to one another and the reference using MUSCLE, generating a FASTA with gaps in the alignment represented by -.
flowchart TD
A[sample 1 consensus] & B[sample 2 consensus] & C[sample 3 consensus] & D[reference FASTA]---q{{multiple sequence alignment}}--->E[multiple alignment FASTA]
[1] Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter ML. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009 Jul;19(7):1195-201. doi: 10.1101/gr.091231.109. Epub 2009 May 13. PMID: 19439516; PMCID: PMC2704435.