Welcome to the project page of DeepVesselGraph A Dataset and Benchmark for Graph Learning and Neuroscience.
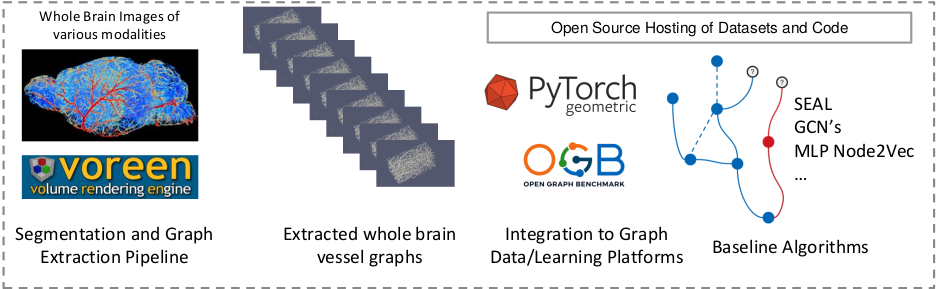
Biological neural networks define human and mammalian brain function and intelligence and form ultra-large, spatial, structured graphs. Their neuronal organization is closely interconnected with the spatial organization of the brain's microvasculature, which supplies oxygen to the neurons and builds a complementary spatial graph. In this project we are providing an extendable dataset of whole-brain vessel graphs based on various multi-center imaging protocols.
This new dataset paves a pathway towards translating advanced graph learning research into the field of neuroscience. Complementarily, the new dataset raises challenging graph learning research questions for the machine learning community, for example how to incorporate biological priors in a meaningful and interpretable way into learning algorithms.
- Whole brain vessel graphs: are the key for research questions to Biology and Neuroscience, including Neuronal organisation, stroke modeling and hemodynamics
- Ready-to use and large set of data: We are providing whole brain graphs from different research groups and will continously update our dataset.
- Data-Loaders: We are providing extensive functions to readily process our data for machine learning research, including the community standard OGB and pyG dataloaders
- Benchmarks: We benchmarked a comprehensive set of state of the art methods in link prediction and node classification; we provide all codes and detailed instructions
- Open-source, “living” initiative: VesselGraph is an open source initiative. We want to expand our datasets as soon as other brain imaging becomes publicly available
Authors Authors Authors "Whole Brain Vessel Graphs: A Dataset and Benchmark for Graph Learning and Neuroscience (VesselGraph)", arXiv:XYZ
Please cite this work if any of our code or datasets are helpful for your research. Considering the specific graphs, please also cite the respective original datasets as described in the preprint.
Our software is licensed under the MIT license. The data is licensed under a Creative Commons Namensnennung-Nicht kommerziell 4.0 International Lizenz.
We provide our graphs as preprocessed OGB datasets (OGBN and OGBL) that are automatically retrieved by the dataloaders when executing the algorithms in ./source/baseline_models/.
For this step, please run ./source/ogb_dataset/link_prediction/update_ogbl_master.sh and ./source/ogb_dataset/node_classification/update_ogbn_master.sh once, before you execute any
of the algorithms in ./source/vaseline_models..
If you would rather work with customized solutions (different datasplits, etc.), we provide you with all steps of our pipeline to generate, preprocess and convert the raw graphs to PyG and OGB formats. In the following section, we describe how our graphs have been built. You are invited to skip this section if you prefer working with our preprocessed graphs.
Use Voreen Graph Generation Tool to make the node_list and edge_list from a segmentation volume.
Go to ./source/dataset_preprocessing/ and run process_edge_list.py with arguments of --node_list and --edge_list
| Dataset Name | Unprocessed | Preprocessed |
|---|---|---|
| BALBc_no1 | download | download |
| BALBc_no2 | download | download |
| BALBc_no3 | download | download |
| C57BL_6_no1 | download | download |
| C57BL_6_no2 | download | download |
| C57BL_6_no3 | download | download |
| CD1-E_no1 | download | download |
| CD1-E_no2 | download | download |
| CD1-E_no3 | download | download |
Got to ./source/feature_generation/atlas_annotation/ and run generate_node_atlas_labels.py with arguments of --node_list and --edge_list
Got to ./source/pytorch_dataset/ and run link_dataset.py and node_dataset.py to create pytorch-geometric compatible dataset for link-prediction and node-classification task.
-
For Graph
$\mathcal{G}$
-
RANDOM SEED: the seed can be fixed in
./source/ogb_dataset/link_prediction/python3 generate_ogbl_dataset.py -
DATA SPLIT: The train/validation/test split can be set using the function
./source/ogb_dataset/link_prediction/vesap_utils.py -
Got to
./source/ogb_dataset/link_prediction/and runpython3 generate_ogbl_dataset.pywith argument--datasetfrom the list ofDataset Namein the table above and--splitting_strategyeitherrandomorspatial -
Subsequently run
update_ogbl_master.shfor compiling the ogb repository locally.
-
For Line Graph
$L(\mathcal{G})$
-
RANDOM SEED: the seed can be fixed in
./source/ogb_dataset/node_prediction/generate_ogbn_dataset.py -
DATA SPLIT: The train/validation/test split can be set by modifying
./source/ogb_dataset/node_prediction/generate_ogbn_dataset.py -
Go to
./source/ogb_dataset/node_classification/and runpython3 generate_ogbl_dataset.pywith argument--datasetfrom the list ofDataset Namein the table above -
Subsequently run
update_ogbn_master.shfor compiling the ogb repository locally. -
We use the following options
Enter indices of desired features (Use "," to separate them): 0,1,2
Enter feature index of desired label: 4
Choose between a certain number of balanced classes (bc) or define classes by pixel boundaries (pb): pb
Enter desired radius boundaries as pixel values (Use "," to separate them): 5,13.33- Subsequently run
update_ogbn_master.shfor node-classification task.
We provide PyG dataset classes for link and node prediction tasks in source/pytorch_dataset/. Utilize LinkVesselGraph and NodeVesselGraph respectively.
We store our graphs as OGBN (OGB Node Prediction) and OGBL (Link Prediction) graphs. All algorithms in source/baseline_models/ rely on OGB Dataloaders
and process the graphs in OGB compatible format.
- Training
Go to ./VesselGraph/source/baseline_models/link_prediction/ and select a MODEL directory to run
e.g. to run GNN, one needs to use the following python3 gnn.py --dataset DATASET_NAME.
For the dataset name, we follow the OGB convention. For example, to run the BALBc_no1 whole brain with a spatial splitting strategy, and without edge features (edge attributes),
use python3 gnn.py --dataset ogbl-BALBc_no1_spatial_no_edge_attr.
We also provide a memory-friendly alternative (a selected Region of Interest of the entire graph). To run the models on the selected region of interest,
use python3 gnn.py --dataset ogbl-link_vessap_roi3_spatial_no_edge_attr.
If you are unsure what options are available, simply run the following command that will list all available datasets:
use python3 gnn.py --dataset ogbl-xyz.
Download Trained Weight
| Model Name | Checkpoint |
|---|---|
| Adamic Adar | [heuristic, not applicable] |
| Common Neighbors | [heuristic, not applicable] |
| Resource Allocation | [heuristic, not applicable] |
| Matrix Factorization | download |
| MLP | download |
| GCN GCN | download |
| GCN GCN + embeddings | download |
| GCN SAGE + embeddings | download |
| GCN SAGE | download |
| SEAL | download |
- Testing
Go to ./VesselGraph/source/baseline_models/link_prediction/ and select go a MODEL directory to run
e.g. to run GNN, one needs to use the following python3 gnn.py --test_only --dataset DATASET_NAME
For the dataset name, we follow the OGB convention. For example, to run the BALBc_no1 whole brain with minimum radius labels,
use python3 gnn.py --dataset ogbn-BALBc_no1_pb_minRadiusAvg.
We also provide a memory-friendly alternative (a selected Region of Interest of the entire graph). To run the models on the selected region of interest,
use python3 gnn.py --dataset ogbn-node_vessap_roi3_pb_minRadiusAvg.
If you are unsure what options are available, simply run the following command that will list all available datasets:
use python3 gnn.py --dataset ogbn-xyz.
- Training
Download Trained Weight
| Model Name | Checkpoint |
|---|---|
| GCN | download |
| GraphSAGE | download |
| GraphSAINT | download |
| SIGN | download |
| Cluster-GCN | download |
| MLP | download |
| SpecMLP-W + C&S | download |
| SpecMLP-W + C&S + N2Vec | download |
- Testing
We are a living and continously maintained repository! Therefore, we welcome contributions of additional datasets and methods! There are multiple ways to contribute; if you are willing to share whole brain segmentations and graphs ....