GenMap is a tool to compute the mappability respectively frequency of nucleotide sequences (DNA and RNA). In particular, it computes the (k,e)-frequency, i.e., how often each k-mer from the sequence occurs with up to e errors in the sequence itself. The (k,e)-mappability is the inverse of the (k,e)-frequency. Hence, a mappability value of 1 at position i indicates that the k-mer in the sequence at position i occurs only once in the sequence with up to e errors. A low mappability value indicates that this k-mer belongs to a repetitive region.
A small example on how to run GenMap is listed below, for detailed examples such as marker sequence computation on multiple fasta files, please check out our GitHub Wiki pages.
For questions or feature requests feel free to open an issue on GitHub or send an e-mail to
christopher.pockrandt [ÄT] fu-berlin.de.
Christopher Pockrandt, Mai Alzamel, Costas S. Iliopoulos, Knut Reinert. GenMap: Fast and Exact Computation of Genome Mappability. bioRxiv, presented on RECOMB-Seq, 2019.
Your CPU must support the POPCNT instruction.
If you have a modern CPU, you can go with the optimized 64 bit version that additionally uses up to SSE4 (MMX, SSE, SSE2, SSE3, SSSE3, SSE4).
This improves the running time by 10 %.
To verify whether your CPU supports these instructions sets you can check the output of
cat /proc/cpuinfo | grep -E "mmx|sse|popcnt" (Linux) or
sysctl -a | grep -i -E "mmx|sse|popcnt" (Mac).
| Platform | Download | Version | Additional requirements |
| Linux 64 bit | 1.0.2 (2019-09-04) | - | |
| Linux 64 bit optimized | 1.0.2 (2019-09-04) | requires up to SSE4 | |
| Mac 64 bit | 1.0.2 (2019-09-04) | - | |
| Mac 64 bit optimized | 1.0.2 (2019-09-04) | requires up to SSE4 |
Please note that building from source can easily take 10 minutes and longer depending on your machine and compiler.
$ git clone --recursive https://github.com/cpockrandt/genmap.git $ mkdir genmap-build && cd genmap-build $ cmake ../genmap -DCMAKE_BUILD_TYPE=Release $ make genmap $ ./bin/genmap
If you are using a very old version of Git (< 1.6.5) the flag --recursive does not exist.
In this case you need to clone the submodule separately before you can run cmake:
$ git clone https://github.com/cpockrandt/genmap.git $ cd genmap $ git submodule update --init --recursive
- Operating System
- GNU/Linux, Mac
- Architecture
- Intel/AMD platforms that support
POPCNT - Compiler
- GCC ≥ 4.9, LLVM/Clang ≥ 3.8
- Build system
- CMake ≥ 3.0
- Language support
- C++14
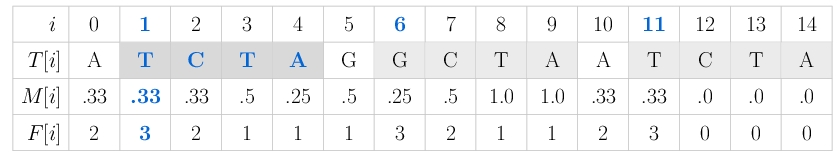
Below you can see the (4,1)-mappability and frequency M and F of the nucleotide sequence T = ATCTAGGCTAATCTA.
Only mismatches (Hamming distance) are considered.
The mappability value M[1] = 0.33 means that the 4-mer starting at position 1 T[1..3] = TCTA occurs three times in the sequence with up to one mismatch, namely at positions 1 (TCTA), 6 (GCTA) and 11 (TCTA).
The mappability can be exported in various formats that allow post-processing or display in genome browsers.
At first you have to build an index of the fasta file(s) whose mappability you want to compute. This step only has to performed once. You might want to check out prebuilt indices for download.
$ ./genmap index -F /path/to/fasta.fasta -I /path/to/index/folder
A new folder /path/to/index/folder will be created to store the index and all associated files.
There are two algorithms that can be chosen for index construction.
One uses RAM (radix), one uses secondary memory (skew).
Depending on the quota and main memory limitations you can choose the appropriate algorithm with -A radix or
-A skew.
For skew you can change the location of the temp directory via the environment variable (e.g., to choose a directory
with more quota):
$ export TMPDIR=/somewhere/else/with/more/space
To compute the (30,2)-mappability of the previously indexed genome, simply run:
$ ./genmap map -E 2 -K 30 -I /path/to/index/folder -O /path/to/output/folder -t -w -b
This will create a text, wig and bed file in /path/to/output/folder storing the computed mappability in
different formats. You can formats that are not required by omitting the corresponding flags -t -w or -b.
Instead of the mappability, the frequency can be outputted, you only have to add the flag -fl to the previous
command.
A detailed list of arguments and explanations can be retrieved with --help:
$ ./genmap --help $ ./genmap index --help $ ./genmap map --help
More detailed examples can be found in the Wiki.
Building an index on a large genome takes some time and requires a lot of space. Hence, we provide indexed genomes for download.
If you need other genomes indexed and do not have the computational resources, please send an e-mail to christopher.pockrandt [ÄT] fu-berlin.de.
| Genome | Index size (compressed) | Download |
| Human GRCh38 (hg38 patch 13) | 6.6 GB | GRCh38 index |
| Human GRCh37 (hg19 patch 13) | 6.4 GB | GRCh37 index |
| Mouse GRCm38 (mm10 patch 6) | 5.7 GB | GRCm38 index |
| Fruitfly D. melanogaster (dm6 rel. 6) | 0.3 GB | dm6 index |
| Worm C. elegans (ce11 WBcel235) | 0.2 GB | ce11 index |