Please cite our paper when using this open-source module or the AutoClass method:
Li H, Brouwer CR, Luo W. A Universal Deep Neural Network for In-Depth Cleaning of Single-Cell RNA-Seq Data. Nature Communications, 2022, 13:1901, https://doi.org/10.1038/s41467-022-29576-y
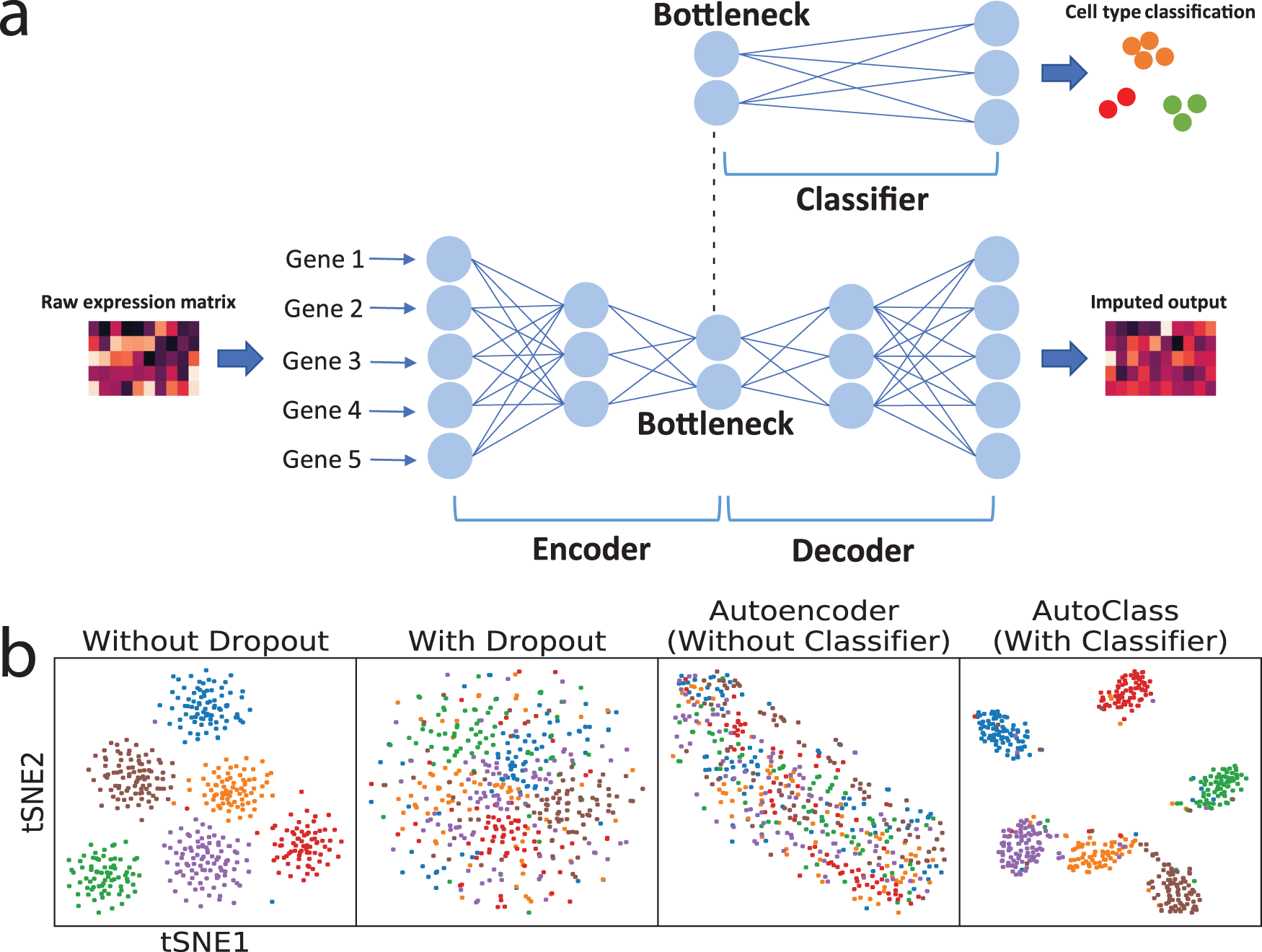
AutoClass is a deep learning tool for scRNA-Seq data cleaning and denoising. AutoClass integrates two deep neural network components, an autoencoder and a classifier, as to maximize both noise removal and signal retention. AutoClass has 3 important features:
- AutoClass is free of distribution assumptions, and effectively cleans a wide range of noises and artifacts.
- AutoClass outperforms the state-of-art methods in multiple types of scRNA-Seq data analyses: i.e. data recovery, differential expression, clustering analysis and batch effect removal.
- AutoClass is robust on key hyperparameter settings: i.e. bottleneck layer size, pre-clustering number and classifier weight.
Fig. 1: AutoClass integrates a classifier to a regular autoencoder, as to fully reconstruct scRNA-Seq data.
You can download AutoClass module from GitHub. AutoClass runs with Python 3, and you need to have TensorFlow (>=2.0) and a few other pacakges installed first(as below).
pip install --upgrade tensorflow
pip install numpy pandas sklearn matplotlibDownload or clone AutoClass module (replace with your own local directory):
cd /path/to/your/local/directory/
git clone https://github.com/datapplab/AutoClassSnapshot of the initial release:
In this repository, you can find several tutorials on AutoClass with Full examples.
- Introductory tutorial
- Example analysis with simulated scRNA-Seq data (Dataset 1 in the paper)
- Example analysis with real scRNA-Seq data (Baron Dataset in the paper)
- Denoising multiple types of noise beyond dropout
- Denoising and clustering of multiple real scRNA-Seq datasets
Demo datasets used in these tutorials can be found in the datasets directory.
Please check the tutorials, module documentation and the AutoClass paper for more info. You can reach the author at hli15[AT]tulane.edu or luo_weijun[AT]yahoo.com.
Thank you for your interest.