Emil Westin
This document contains notes I make about R, data science and software engineering in the context of statistics.
How to use this document: search for a keyword with Ctrl + f or
scroll trough.
The following code assumes you are using windows and can be run from
RStudio. The version of R can usually be found here
C:\Program Files\R\R-4.2.0. More
info.
install.packages("installr")
library(installr)
# Run the following
# (you can also just type installr::updateR() and choose options interactively)
installr::updateR(browse_news = T,
fast = F,
install_R = T,
copy_packages = T,
copy_site_files = T,
keep_old_packages = T,
update_packages = F, # this will be done in next step
start_new_R = F,
quit_R = F
)
# When the above is completed, restart R, exit R studio and reopen with a fresh session
# Reinstall the copied packages to match the latest version of R
update.packages(checkBuilt=TRUE, ask=FALSE)
# Double check that the packages have been built with the latest version of R:
installed.packages()# unloading library(officer)
detach(package:officer)| Command | |
|---|---|
| Restart R-session and unload packages: | Command/Ctrl + Shift + F10 |
| Comment/uncomment single/multiple lines: | Ctrl+ Shift + c |
| Display command palette: | Ctrl+ Shift + p |
| Auto-indent: | Ctrl + i |
| Edit multiple lines simultaneusly (increase the blinking line vertically): | Ctrl + Alt + up / down |
| Duplicate current line: | Alt + Shift + up / down |
Insert assignment (<-): |
Alt + - |
Insert pipe operator (%>%): |
Ctrl + Shift + m |
| Delete current line: | Ctrl + d |
| Multiple word selection: | Ctrl + Alt + double left click |
| Run code from beginning to line | Ctrl + Alt + b |
| Move/shift line up/down | Alt + up / down |
Note: for this to work, the file obviously needs to exist beforehand. If needed, create an empty workbook beforehand:
wb <- createWorkbook(title = str_glue("Månadsuppdatering {upp_datum}"))
addWorksheet(wb, "Session_info")
addWorksheet(wb, "Log")
saveWorkbook(wb, file = path_to_file, overwrite = TRUE)Now if we want to update the ‘Log’-sheet only and not overwrite anything else:
library(openxlsx)
library(tidyverse) # (optional, for using tibble etc)
wb <- openxlsx::loadWorkbook(path_to_file)
df <- openxlsx::readWorkbook(wb, sheet = "Log")
df <- as_tibble(df)
# * do stuff to df*
# write data to the same sheet
openxlsx::writeData(wb = wb, sheet = "Log", x = df)
# save it:
openxlsx::saveWorkbook(wb, path_to_file, overwrite = TRUE)This is the absolutely best method if you want to add more sheets as time goes and gives the most flexibility. It also preserves the original formatting of the file that is loaded and allows you to add any styling you want at any time.
If you’re just interested in reading the excel data without manipulating it too much (when saving it), these are some of the simplest options.
# Read xlsx/xls:
readxl::read_xlsx(name_of_file)
readxl::read_xls(name_of_file) # or readxl::read_excel(name_of_file) for guessing xls or xlsx
# Read xlsx:
openxlsx::read.xlsx(name_of_file) # only for xlsx
# The following library is the best for ensuring the encoding is correct.
# Especially when the columns contain messy data, maybe there is is a mix of , or . in a number column.
# This function will ensure it is read exactly as it is written in the excel-file. Requires name or index of sheet.
library(xlsx)
xlsx::read.xlsx(name_of_file, sheetIndex = 1, encoding = "UTF-8")
# Custom function useful when used for reading multiple excel files in purrr::map()
read_my_excel <- function(x) {
print(str_glue("Reading {x}"))
# try(openxlsx::read.xlsx(x, sep.names = " ") %>%
# as_tibble())
try(xlsx::read.xlsx(x, sheetIndex = 1, encoding = "UTF-8") %>%
as_tibble())
}
# something like this: (saved as df)
excel_df <- list.files(dir_where_excel_files_are, full.names = T) %>%
set_names(~basename(.) %>% str_remove(".xlsx")) %>%
enframe(name = "filename") %>%
mutate(excel_in = map(value, read_my_excel))
# Password protected excel:
library("excel.link")
xl.read.file(indata, password = "mypswd", write.res.password="mypswd")
# Write excel files:
library(openxlsx)
write.xlsx(df, file = "Output/myfile.xlsx")Option 1 (simplest):
# Write multiple data frames as sheets in single excel file:
list_of_datasets <- list("Sheet name 1" = df1,
"Sheet name 2" = df2,
"Sheet name 3" = df3)
openxlsx::write.xlsx(list_of_datasets, file = "Output/myfile.xlsx") Option 2 (more customization):
library(openxlsx)
library(tidyverse)
# Split dataframe into list of tables
dat <- split(mtcars, mtcars$cyl)
wb <- createWorkbook()
# loop trough list of splitted tables, adding worksheets and styles:
map2(.x = dat, .y = names(dat),
.f = function(data, name) {
addWorksheet(wb, name)
freezePane(wb, name, firstActiveRow = 2)
writeData(wb, name, data, withFilter = TRUE)
})
saveWorkbook(wb, file = "Output/myfile.xlsx", overwrite = TRUE)If you don’t need to apply any styling for each sheet you can just do:
dat <- split(mtcars, mtcars$cyl)
openxlsx::write.xlsx(dat, file = "Output/myfile.xlsx") comments <- c("comment 1",
"comment 2",
"comment 3")
# add comments to columns:
map(1:length(comments),
~ writeComment(wb, 1, col = .x, row = 1,
comment = createComment(comment = comments[.x], visible = F)))
# add comments to rows:
map(1:length(comments),
~ writeComment(wb, 1, col = 1, row = .x,
comment = createComment(comment = comments[.x], visible = F)))# header style
hs1 <- openxlsx::createStyle(
fontColour = "#ffffff", fgFill = "#4F80BD",
halign = "center", valign = "center", textDecoration = "bold",
border = "TopBottomLeftRight", fontSize = 14
)
curr_sheetname <- str_glue("<insert_sheetname_here>")
wb <- openxlsx::createWorkbook()
openxlsx::addWorksheet(wb, curr_sheetname)
# If you want to change base font:
openxlsx::modifyBaseFont(wb, fontSize = 12, fontColour = "black", fontName = "Arial Narrow")
# Write data
openxlsx::writeData(wb, sheet = curr_sheetname, data, withFilter = T, headerStyle = hs1)
openxlsx::freezePane(wb, sheet = curr_sheetname, firstRow = T)
openxlsx::setColWidths(wb, 1, cols = 1:ncol(data), widths = "auto")
# Optional styling
# Add bgcolor based on cell values. Based on min/max when rule=NULL
conditionalFormatting(wb,
curr_sheetname,
cols = ncol(data)-3,
rows = 2:(nrow(data)+1),
style = c("lightblue", "darkred"),
rule = NULL,
type = "colourScale"
)
# Change format of columns
openxlsx::addStyle(wb, sheet = curr_sheetname, createStyle(numFmt = "NUMBER"), rows = 2:nrow(tmp), cols = 1)
# map(cols_to_format,
# ~openxlsx::addStyle(wb, sheet = curr_enkät, createStyle(numFmt = "NUMBER"), rows = 2:nrow(tmp), cols = .x))
openxlsx::saveWorkbook(wb, file = str_glue("Output/insert_name.xlsx"), overwrite = T)
# For multiple sheets at once:
wb <- openxlsx::createWorkbook()
openxlsx::modifyBaseFont(wb, fontSize = 11, fontColour = "black", fontName = "Arial")
sheets <- df$respondentkategori |> unique()
map(sheets, ~openxlsx::addWorksheet(wb, .x))
map(sheets, ~openxlsx::writeData(wb, sheet = .x, df |> filter(respondentkategori==.x), withFilter = T, headerStyle = hs1))
map(sheets, ~openxlsx::freezePane(wb, sheet = .x, firstRow = T))
map(sheets, ~openxlsx::setColWidths(wb, sheet = .x, cols = 1:ncol(df |> filter(respondentkategori==.x)), widths = "auto"))
openxlsx::saveWorkbook(wb, file = str_glue("Output/insert_name.xlsx"), overwrite = T)counter <- 1
for (i in seq_along(x)) {
startindex <- counter
tmp <- x[[i]]
n <- nrow(tmp)+1 # antal rader + en rad för kolumnen
title <- str_remove(names(x[i]), "X") # enkätfrågan + ta bort X
openxlsx::writeData(wb, curr_enkät, title, startRow = startindex, startCol = 1)
openxlsx::addStyle(wb, curr_enkät, style, rows = startindex, cols = 1)
openxlsx::writeDataTable(wb, curr_enkät, tmp, startRow = startindex+1, startCol = 1)
counter <- counter + n + 1 + 1
rm(tmp)
rm(title)
rm(startindex)
rm(n)
}When importing dates from Excel into R, dates are represented as days since 1899-12-30. Usually this is done automatically, but in some cases there may be text in the columns, then you can use this method (text -> NA):
as.Date(44301, origin = "1899-12-30")All versions of Excel for Windows calculate dates based on the 1900 date system. However for older versions of Mac, the 1904 date system is needed. So it is a good idea to double check with the data so that your imported dates look as expected. Link
If the column contains both dates and text for some reason, and you want to keep the text, use this function:
# Function for converting excel-dates in 1900-date system to R-dates, while keeping texts as they are
# if the column has a mix of texts and dates
convertd <- function(x) {
x <- ifelse(str_detect(x, "^[0-9]+$"), as.character(as.Date(as.numeric(x), origin = "1899-12-30")), x)
return(x)
}
convertd(c(22,"aa"))
# [1] "1900-01-21" "aa" mergemycells <- function(df, wb, sheet, column, offset = 1, .by=NULL, filtrera = TRUE) {
# in this case we filter the dataframe by a variable corresponding to the sheet name
if (filtrera) {
tmp <- df |> filter(respondentkategori==sheet)
} else {
tmp <- df
}
#offset <- 1 # om rader/kolumner börjar på rad/kolumn 2 ist för 1
# if by is not null, the merging will be performed on by but will use column as reference for which ids to target
if (!is.null(.by)) {
# dep <- deparse(substitute(.by))
dep <- .by
} else {
dep <- deparse(substitute(column))
}
whichcol <- which(names(tmp) == dep)+offset
# get unique values from column name
unika <- tmp |> pull({{column}}) |> unique()
# check which row ids match with unique values above
rader <- map(1:length(unika), ~which(tmp |> pull({{ column }}) %in% unika[.x])+(offset+1))
#return(rader)
# split again if the categories will repeat further on
# it will return a list of unbroken sequences
rader <- map(rader, ~ split(.x, cumsum(c(TRUE, diff(.x)!=1)))) |> flatten()
# merge cells
map(rader, ~mergeCells(wb = wb, sheet = sheet, cols = whichcol, rows = .x))
# add style for merged cells
map(rader, ~addStyle(wb = wb, sheet = sheet, style = createStyle(valign = "center", halign = "center"), cols = whichcol, rows = .x, stack = T))
}
# merge column 'Hej' based on unbroken rows
map(sheets, ~ mergemycells(svar1, wb, .x, column=Hej))
# område will merge cells based on the same rows as the column Hej
map(sheets, ~ mergemycells(svar1, wb, .x, column=Hej, .by= "Område" ))mycolors <- list("Grön" = createStyle(fgFill = "#23FF00"),
"Gul" = createStyle(fgFill = "#FFFB00"),
"Röd" = createStyle(fgFill = "#FF0000")
)
conditional_colors_by_text2 <- function(df, wb, sheets, colors, target_cols) {
for (i in target_cols) {
for (.color in names(colors)) {
map(.x = sheets,
~ addStyle(wb = wb,
sheet = .x,
style = mycolors[[.color]],
rows = which(grepl(df |> select(all_of(i)) |> pull(), pattern = .color))+1,
cols = which(names(df) %in% i)
)
)
}
}
}
conditional_colors_by_text2(farger, wb, c("sheet1", "sheets2"), mycolors, target_cols = c("col1", "col2"))Credits to Luke C on StackOverflow.
OutsideBorders <-
function(wb_,
sheet_,
rows_,
cols_,
border_col = "black",
border_thickness = "medium") {
left_col = min(cols_)
right_col = max(cols_)
top_row = min(rows_)
bottom_row = max(rows_)
# https://stackoverflow.com/questions/54322814/how-to-apply-thick-border-around-a-cell-range-using-the-openxlsx-package-in-r
sub_rows <- list(c(bottom_row:top_row),
c(bottom_row:top_row),
top_row,
bottom_row)
sub_cols <- list(left_col,
right_col,
c(left_col:right_col),
c(left_col:right_col))
directions <- list("Left", "Right", "Top", "Bottom")
mapply(function(r_, c_, d) {
temp_style <- createStyle(border = d,
borderColour = border_col,
borderStyle = border_thickness)
addStyle(
wb_,
sheet_,
style = temp_style,
rows = r_,
cols = c_,
gridExpand = TRUE,
stack = TRUE
)
}, sub_rows, sub_cols, directions)
}
map(sheets,
~ OutsideBorders(
wb,
sheet_ = .x,
rows_ = 0:nrow(svar1)+1,
cols_ = 0:ncol(svar1)
)
)
# Optional: also add borders inside:
kanter <- createStyle(border = "TopBottomLeftRight", borderStyle = "thin", borderColour="black", wrapText = TRUE, halign = "left", valign = "top")
# important to use stack=TRUE
map(sheets,
~openxlsx::addStyle(wb, .x, kanter,
rows = 1:(nrow(svar1)+1),
cols = 1:(ncol(svar1)), gridExpand = T,
stack = T))Will hide columns but can easily be expanded.
hidecols <- c("col1", "col2") # specify which column names to hide
map(sheets, ~groupColumns(wb, sheet = .x, cols = which(names(svar1) %in% hidecols), hidden = T))ss <- "my sheet"
startcol <- 4
startrow <- 17
# fill xy-coordinated with vales from another sheet
openxlsx::writeFormula(wb, ss, x = "=AnotherSheet!U3" ,xy = c(startcol, startrow))
# writedata can also be used to write to specific cells
openxlsx::writeData(wb, ss, 9999, xy = c(startcol, startrow+1))
openxlsx::writeData(wb, ss, 12, xy = c(startcol, startrow+2))library(tidyverse)
library(openxlsx)
wb <- createWorkbook()
addWorksheet(wb, "Sheet 1", gridLines = FALSE)
p <- iris %>%
ggplot(aes(x=Sepal.Length, y = Sepal.Width)) +
geom_point()
print(p)
insertPlot(wb, sheet = "Sheet 1", width = 20, height = 20, fileType = "png", units = "cm")
## Save workbook
saveWorkbook(wb, "insertPlotExample2.xlsx", overwrite = TRUE)library(tidyverse)
library(openxlsx)
wb <- createWorkbook()
addWorksheet(wb, "Sheet 1", gridLines = FALSE)
ccc <- iris$Species %>% unique
startrow <- 1
for (i in ccc) {
#print(i)
p <- iris %>%
filter(Species == i) %>%
ggplot(aes(x=Sepal.Length, y = Sepal.Width)) +
geom_point()
print(p)
insertPlot(wb, sheet = "Sheet 1", startRow = startrow, width = 20, height = 20, fileType = "png", units = "cm")
startrow <- startrow + 45
}
getwd()
## Save workbook
saveWorkbook(wb, "insertPlotExample2.xlsx", overwrite = TRUE)Credits to https://dominicroye.github.io/en/2019/import-excel-sheets-with-r/
in_path <- "path/to/excelfile.xlsx"
# read multiple sheets and join them into single dataframe:
df <- in_path %>%
readxl::excel_sheets() %>% # vector containing name of sheets
set_names() %>% # sets the names of vector with names of sheets
# map_df: a list of tables joined with bind_rows into single dataframe:
purrr::map_df(readxl::read_excel,
path = in_path,
.id = "level" # creates column called level with names of the sheets
)read_multiple_excel_sheets <- function(path, idname = "sheet") {
print(str_glue("Reading {path} with id = {idname} for each sheet"))
path %>%
readxl::excel_sheets() %>%
set_names() %>%
map_df(read_excel, path = path, .id = idname)
}
df <- dir(pattern = "xlsx") %>%
map_df(read_multiple_excel,
.id = "level")Read multiple csv files in directory using summarise.
See https://www.tidyverse.org/blog/2020/03/dplyr-1-0-0-summarise/
library(tidyverse)
# reading all csv files in current directory:
tibble(path = dir(pattern = "\\.csv$")) %>%
rowwise(path) %>%
summarise(read_csv(path))Or read from any directory:
path <- "C:/Users/my_usrname/output/csv"
df <- list.files(path, full.names = T, include.dirs = F) %>%
set_names(basename(.)) %>%
map_df(.x = .,
.f = read_delim,
delim = ";",
col_types = cols(.default = "c"),
.id = "matris")If the csv files has multiple columns with same names, try to merge them together. For example, all columns starting with ‘SOURCE’:
# get colnames
source_cols <- res[str_detect(names(res), "SOURCE.*")] %>% names()
x <- res[source_cols]
res$SOURCE <- do.call(paste, c(x, sep = " # ")) # paste all cols together, separate with # in this case
res <- res %>% select(-setdiff(source_cols, "SOURCE")) # remove all other columns This is useful, especially since csv files may in fact be separated with semicolon (;) and not comma, which may be due to the locale and OS used when the csv file was saved. For example, in Swedish, comma (,) is used as a decimal separator.
res <- list.files(path = indir, pattern = "*.csv", full.names = TRUE) %>%
map_df(read_delim,
delim = ";",
col_types = cols(.default = "c"), # read all cols as chr
locale = locale(encoding = "iso-8859-15"), # specify locale if needed, in this case ISO Latin 9 (western europe)
id="filename")library(flextable) # converting dataframes to flextable objects
library(officer) # for adding data to word document, like flextables
# simplest example:
tab1 <- matrix( c(1,2,3,4), ncol=2, nrow=2)
word_export <- read_docx()
word_export <- word_export %>% body_add_flextable( as.data.frame.matrix(tab1) %>% flextable() )
print(word_export, 'try.docx')# add new page:
mydoc %>% body_add_break()Function for adding R table to word document:
myft <- function(mydoc, tab, title) {
res <- body_add_par(mydoc, "")
res <- body_add_par(mydoc, title, style = "Tabellrubrik_")
res <- body_add_flextable(mydoc, flextable(tab %>% as.data.frame.matrix() %>% rownames_to_column(" ")) %>% autofit(), align = "left")
return(res)
}
mydoc <- myft(mydoc, tt0, "Table 1. xxx")library(flextable)
library(officer)
setwd("<set working dir>")
getwd()
inmall <- "word_template.docx"
utmall <- "out_file.docx"
# RAPPORT: IMPORT --------------------------------------------------------------
mydoc <- read_docx(inmall)
# RAPPORT: TITLE PAGE ----------------------------------------------------------
mydoc <- body_replace_all_text(mydoc, "TITLE", "My title page", only_at_cursor = F)
mydoc <- body_replace_all_text(mydoc, "DATE", as.character(Sys.Date()), only_at_cursor = F)
# RAPPORT: SECTION 1 -----------------------------------------------------------
mydoc <- body_add_par(mydoc, "Descriptive statistics", style = "heading 1", pos = "before")
# RAPPORT: EXPORT --------------------------------------------------------------
print(mydoc, utmall)
shell.exec(utmall)To set a reference category for a categorical variable in regression models, you move that category to the first position of the levels. If you want “Mid” to be the reference and the levels are “Low”, “Mid”, “High”, you simply want to change the order of levels to “Mid”, “Low”, “High”.
# relevel single variable
df %>%
dplyr::mutate(q_mycat = fct_relevel(q_mycat, "Mid"))
# explicitly same as:
df %>%
dplyr::mutate(q_mycat = factor(q_mycat, c("Mid", "Low", "High")))
# using stats::relevel:
df$q_mycat <- relevel(df$q_mycat, ref = "Mid")
# relevel multiple variables (assuming they have the same factors levels):
df %>%
dplyr::mutate(across(starts_with("q_"), ~ fct_relevel(.x, "Mid")))library(tidyverse)
map_df(iris, ~sum(is.na(.))) %>% pivot_longer(everything(), names_to = "Variable", values_to = "n")# A tibble: 5 × 2
Variable n
<chr> <int>
1 Sepal.Length 0
2 Sepal.Width 0
3 Petal.Length 0
4 Petal.Width 0
5 Species 0
library(tidyverse)
get_summary <- function(.data) {
.data %>%
mutate(across(where(is.factor), as.character)) %>%
summarise(across(everything(),
.fns = list(n_missing = ~sum(is.na(.)),
minimum = ~min(., na.rm = T),
maximum = ~max(., na.rm = T),
mean = ~ ifelse(is.numeric(.), mean(., na.rm = T), NA),
median = ~ ifelse(is.numeric(.), median(., na.rm = T), NA)
# minimum = ~ ifelse(is.numeric(.), min(.), NA)
),
.names = "{.col}__{.fn}")) %>%
mutate(across(everything(), as.character)) %>%
pivot_longer(everything(), names_to = "v") %>%
separate(v, into = c("v","t"), sep = "__") %>%
pivot_wider(names_from = t, values_from = value) %>%
mutate(across(c(n_missing, mean, median), as.numeric)) %>%
dplyr::rename("variable" = v)
}
iris %>% get_summary()
# # A tibble: 5 × 6
# variable n_missing minimum maximum mean median
# <chr> <dbl> <chr> <chr> <dbl> <dbl>
# 1 Sepal.Length 0 4.3 7.9 5.84 5.8
# 2 Sepal.Width 0 2 4.4 3.06 3
# 3 Petal.Length 0 1 6.9 3.76 4.35
# 4 Petal.Width 0 0.1 2.5 1.20 1.3
# 5 Species 0 setosa virginica NA NA table()
prop.table() # with proportions# 2=sum cols, 1=sum rows, otherwise it sums both
tab <- addmargins(table(df$Company,df$Marital), 2)library(tidyverse)
# Old way (superseded):
iris %>%
mutate_if(is.factor, as.character)
# New way (recommended from version 1.0.0):
iris %>%
mutate(across(where(is.factor), as.character))
# if converting to factor, use as_factor to preserve order that the factors appear
## perform conversion on a list of dataframes
dat <- iris %>% as_tibble() %>% split(.$Species) # list of dataframes split by Species
dat %>%
map(~ .x %>% mutate(across(where(is_double), as.character)))Rename the last column:
bind_rows(sms_tot, mms_tot) %>%
dplyr::rename(value = last_col())
# alternatively if you like to overcomplicate things:
bind_rows(sms_tot, mms_tot) %>%
dplyr::rename(value = !! last(names(sms_tot)))factor_case_when <- function(..., reverse = FALSE) {
args <- as.list(match.call()) # capture args to list
args <- args[-c(1, which(names(args) == "reverse"))] # ...skip first element which is name of func and reverse-arg
lvls <- purrr::map(args, \(x) x[[3]]) |> # use third element
purrr::discard(\(x) is.na(x))
if (reverse) lvls <- rev(lvls)
factor(dplyr::case_when(...), levels = unique(lvls))
}Consider
| A | B |
|---|---|
| 1 | apple, banana, pear |
| 2 | watermelon, apple |
library(tidyverse)
df <- tribble(
~A, ~B,
1, c("apple, banana, pear"),
2, c("watermelon, apple")
)
df %>%
separate_rows(B)# A tibble: 5 × 2
A B
<dbl> <chr>
1 1 apple
2 1 banana
3 1 pear
4 2 watermelon
5 2 apple
Table band_members:
| name | band |
|---|---|
| Mick | Stones |
| John | Beatles |
| Paul | Beatles |
To perform a self-join in R, i.e. a Cartesian product of all
combinations of a table, supply by = character()
library(tidyverse)
band_members %>%
left_join(band_members, by = character())# A tibble: 9 × 4
name.x band.x name.y band.y
<chr> <chr> <chr> <chr>
1 Mick Stones Mick Stones
2 Mick Stones John Beatles
3 Mick Stones Paul Beatles
4 John Beatles Mick Stones
5 John Beatles John Beatles
6 John Beatles Paul Beatles
7 Paul Beatles Mick Stones
8 Paul Beatles John Beatles
9 Paul Beatles Paul Beatles
This is basically the R/dplyr equivalent of a SQL self-join:
select * from band_members b1, band_members b2Table band instruments:
| name | plays |
|---|---|
| John | guitar |
| Paul | bass |
| Keith | guitar |
band_members %>% inner_join(band_instruments)# A tibble: 2 × 3
name band plays
<chr> <chr> <chr>
1 John Beatles guitar
2 Paul Beatles bass
band_members %>% left_join(band_instruments)# A tibble: 3 × 3
name band plays
<chr> <chr> <chr>
1 Mick Stones <NA>
2 John Beatles guitar
3 Paul Beatles bass
band_members %>% right_join(band_instruments)# A tibble: 3 × 3
name band plays
<chr> <chr> <chr>
1 John Beatles guitar
2 Paul Beatles bass
3 Keith <NA> guitar
band_members %>% full_join(band_instruments)# A tibble: 4 × 3
name band plays
<chr> <chr> <chr>
1 Mick Stones <NA>
2 John Beatles guitar
3 Paul Beatles bass
4 Keith <NA> guitar
Turn implicit missing values into explicit missing values. The following function works for data frames in long format where there is a ‘value’ column. If you supply a reference data frame, this function detects which values in the supplied column are missing and adds them with values as NA.
library(tidyverse)
# Function for completing missing values for a column, given a reference df
complete_missing <- function(.x, .reference_df, .colname) {
ax <- deparse(substitute(.colname)) # convert .colname to string
missingl <- setdiff(.reference_df[[ax]], .x[[ax]]) # missing levels
a <- rlang::enquo(.colname) #
.x %>%
mutate(!!a := fct_expand(!!a, missingl)) %>%
complete(!!! syms(setdiff(names(.reference_df), "value")))
}Or if you want to complete a column with a value you specify yourself:
library(tidyverse)
complete_col <- function(.x, .colname, .levels) {
a <- rlang::enquo(.colname)
.x %>%
mutate(!!a := fct_expand(!!a, .levels)) %>%
complete(!!! syms(setdiff(names(.x), "value")))
}Example:
x <- tribble(~Sepal.Length, ~Sepal.Width, ~Petal.Length, ~Petal.Width, ~Species,
1, 2, 3, 4, "setosa"
)
# Convert to long formats:
x_long <- x %>%
pivot_longer(-Species)
iris_long <- iris %>%
pivot_longer(-Species)
x_long# A tibble: 4 × 3
Species name value
<chr> <chr> <dbl>
1 setosa Sepal.Length 1
2 setosa Sepal.Width 2
3 setosa Petal.Length 3
4 setosa Petal.Width 4
x_long %>%
complete_missing(iris_long, Species) %>%
head(10)# A tibble: 10 × 3
Species name value
<fct> <chr> <dbl>
1 setosa Petal.Length 3
2 setosa Petal.Width 4
3 setosa Sepal.Length 1
4 setosa Sepal.Width 2
5 versicolor Petal.Length NA
6 versicolor Petal.Width NA
7 versicolor Sepal.Length NA
8 versicolor Sepal.Width NA
9 virginica Petal.Length NA
10 virginica Petal.Width NA
# Is equivalent to:
# x_long %>%
# mutate(Species = fct_expand(Species, c("versicolor", "virginica"))) %>%
# complete(!!! syms(setdiff(names(iris_long), "value")))From https://stackoverflow.com/questions/14096814/merging-a-lot-of-data-frames
Reduce(function(...) merge(..., all=TRUE), list(df1, df2, df3))Returns list of dataframes.
library(tidyverse)
mtcars %>% split(.$cyl)Returns a dataframe (data.frame) from all combinations of the supplied vectors or factors. Useful if you for ex. want to test a set of hyperparameters without creating nested for loops, just create a dataframe of all combinations and iterate through it.
expand.grid(letters[1:2], 1:3, c("+", "-"))add_columns <- function(df, columns){
new <- rep(NA_character_, length(columns))
names(new) <- columns
mutate(df, !!!new)
}
sourcecols_in_meta <- str_extract(names(meta),"SOURCE.*") %>% na.omit %>% unique()
sourcecols_in_df_final <- str_extract(names(df_final),"SOURCE.*") %>% na.omit %>% unique()
# cols in meta not in df_final:
diffcols <- setdiff(sourcecols_in_meta, sourcecols_in_df_final)
df_final <- df_final %>%
add_columns(diffcols)right = indicating if the intervals should be closed on the right (and
open on the left) or vice versa. include.lowest = indicating if an
x[i] equal to the lowest (or highest, for right = FALSE) “breaks”
value should be included.
cut(tmp2$Antal, breaks = c(-Inf,20,50,Inf), labels = c("\u226420", "21-50", "50-521"))
# will show ≤20 21-50 50-521
cut(0:10, breaks = c(0, 1, 4, 10, Inf), labels = c("0", "1-3", "4-9", "\u226510"), include.lowest = T,right=F )
# [1] 0 1-3 1-3 1-3 4-9 4-9 4-9 4-9 4-9 4-9 ≥10
#Levels: 0 1-3 4-9 ≥10
# <19, ≥20 :
cut(0:20, breaks = c(0, 19, Inf),labels = c("<20", "\u226520"),include.lowest = T,right=T )
#[1] <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 <20 ≥20
#Levels: <20 ≥20From https://statisticsglobe.com/split-vector-into-chunks-in-r
my_vec <- 2008:2021
chunk_length <- 5
split(my_vec,
ceiling(seq_along(my_vec) / chunk_length))$`1`
[1] 2008 2009 2010 2011 2012
$`2`
[1] 2013 2014 2015 2016 2017
$`3`
[1] 2018 2019 2020 2021
From https://statisticsglobe.com/split-vector-into-chunks-in-r
chunk_number <- 2
split(my_vec, # Applying split() function
cut(seq_along(my_vec),
chunk_number,
labels = FALSE))$`1`
[1] 2008 2009 2010 2011 2012 2013 2014
$`2`
[1] 2015 2016 2017 2018 2019 2020 2021
The problem with sum(x, na.rm = TRUE) is that it will return 0 if all categories are NA. Sometimes we want it to be NA, here is how:
sumna <- function(x) {
if(all(is.na(x))) NA else sum(x, na.rm = TRUE)
}library(lubridate)
seq(ymd("2022-01-01"), ymd("2022-12-31"), by = "1 days")
# "2022-01-01" "2022-01-02" "2022-01-03" ... 2022-12-31"
# in base r, replace ymd() with as.Date()mtcars %>% map(., sum) # sum each col in df
pmap_dbl(mtcars, sum) # sum of each row in df
mods <- by_cyl %>% map(~ lm(mpg ~ wt, data = .)) # in each df, create linear modelWhy use map instead of lapply?
-
Syntacically more convenient: You can use model formulas like
~ . + 1instead offunction(x) x + 1 -
Type-specific: you know exactly what type is returned (double, character, dataframe, etc).
-
Nicely integrated with tidyverse
default_cols <- c("Survey", "sub_area", "Insats")
default_df <- default_cols %>% purrr::map_dfc(setNames, object = list(character()))
# A tibble: 0 x 3
# ... with 3 variables: Survey <chr>, sub_area <chr>, Insats <chr>library(tidyverse)
n_iris <- iris %>% group_by(Species) %>% nest()
## A tibble: 3 x 2
## Groups: Species [3]
# Species data
# <fct> <list>
#1 setosa <tibble [50 x 4]>
#2 versicolor <tibble [50 x 4]>
#3 virginica <tibble [50 x 4]>join_df <- function(df_nest, df_other) {
df_all <- inner_join(df_nest, df_other, by = c("name1" = "name2"))
return(df_all)
}
tmp_n2 <- tmp_n %>%
mutate(new_data = map(data, ~ join_df(., df_of_interest)))Consider the dataframe df:
| matrix | filePathIn |
|---|---|
| A | my_dir/A.px |
| B | my_dir/B.px |
| C | my_dir/C.px |
inFile_n <- df %>% group_by(matrix) %>% nest()
# läs in alla matriser:
inFile_alla <- inFile_n %>%
mutate( rrr = map(data, function(x) read.px(x$filePathIn) ) )
# alternatively:
#inFile_n %>%
# mutate( rrr = map(data, ~ read.px(.x$filePathIn) ) )where:
inFile_n:
| matrix <chr> | data <list> |
|---|---|
| A | <tibble [1 x 2]> |
| B | <tibble [1 x 2]> |
| C | <tibble [1 x 2]> |
inFile_alla:
| matrix <chr> | data <list> | rrr <list> |
|---|---|---|
| A | <tibble [1 x 2]> | <tibble [27,000 x 7]> |
| B | <tibble [1 x 2]> | <tibble [25,200 x 6]> |
| C | <tibble [1 x 2]> | <tibble [126,000 x 7]> |
convert_sweref99_to_wgs84 <- function(.x, .y){
# SWEREF 99
sweref99 <- sf::st_sfc(sf::st_point(c(.x, .y), dim = "XY"), crs = 3006)
wgs84 <- sf::st_transform(sweref99, crs = 4326)
sf::st_coordinates(wgs84)
}
convert_sweref99_to_wgs84(6398983, 320011)
# X Y
# 1 61.74469 1.97728See stringr.R for a walk-through.
Based on the first character of each word in a string. Returns a character vector.
library(tidyverse)
generate_varcodes <- function(s) {
str_split(s, boundary("word")) %>%
map(~str_sub(.x, 1,1)) %>%
map(str_to_upper) %>%
map(str_c, collapse = "") %>%
unlist()
}
mpg %>%
select(model) %>%
distinct() %>%
mutate(acronym = generate_varcodes(model)) %>%
head()tj_to_ttoe <- function(tj) {
# For converting Terajoule to Thousand tons of oil equivalent
tj/(0.041868*1000)
}After selecting variables for a table, you can choose to retrieve the data in some of the following ways.
Note: you can also specify “format”: “json” in the query to get the data directly as json.
library(httr)
url <- "https://pxweb.nordicstatistics.org:443/api/v1/en/Nordic Statistics/Geography and climate/Land use/DENS01.px"
url <- URLencode(url) # convert whitespace to ascii %20
query <- '{
"query": [
{
"code": "time",
"selection": {
"filter": "item",
"values": [
"2016",
"2017",
"2018",
"2019"
]
}
}
],
"response": {
"format": "px"
}
}'
r <- POST(url, body=query)
px <- content(r, type="text", encoding = "Windows-1252")
px
# Save as px file:
fileConn <- file("testpx.px")
writeLines(px, fileConn)
close(fileConn)
# Alternatively, load it directly in R by saving to temporary file path
txtPath <- tempfile(fileext = ".px")
writeLines(text = px, con = txtPath)
close(txtPath)
library(pxR)
d <- pxR::read.px(txtPath, encoding = "iso-8859-15") %>% as_tibble()Save query -> Update the query with a fixed starting time point and the new time periods -> Tab delimited with heading.
This may generate something like this:
https://pxweb.nhwstat.org:443/Prod/sq/0978ea50-2b97-4a48-bc58-d4a71806336e.
To see how it was selected, add ?select:
https://pxweb.nhwstat.org:443/Prod/sq/0978ea50-2b97-4a48-bc58-d4a71806336e?select.
To download it, add .relational_table:
https://pxweb.nhwstat.org:443/Prod/sq/0978ea50-2b97-4a48-bc58-d4a71806336e.relational_table
as_tibble(read_delim(
"https://pxweb.nhwstat.org:443/Prod/sq/0978ea50-2b97-4a48-bc58-d4a71806336e.relational_table",
locale = locale(encoding = "latin1"),
delim = "\t" ) )library(pxR)
px_in <- read.px("abcd10.px", encoding='iso-8859-15')library(pxR)
write.px(
px_in,
"abcd10_new.px",
fileEncoding = "iso-8859-15"
)library(pxweb)
pxweb::pxweb_interactive() # interactively browse statistical databases and download px-files via API queriesWhen you want to include both time series on the year of a time series break. For example: 2018, 2019A, 2019B, 2020
add_time_series_breaks <- function(.data) {
.data %>%
pivot_longer(-År, names_to = "namn") %>%
drop_na(value) %>%
mutate(letter = ifelse(str_detect(namn, "fram till"), "A", "B")) %>%
mutate(metod = str_extract(namn, ", [mM]etod.*$|^[mM]etod.*,")) %>%
mutate(namn = str_remove(namn, metod) %>% str_squish()) %>%
distinct() %>%
group_by(År, namn) %>%
mutate(n = n()) %>%
arrange(År, letter) %>%
ungroup() %>%
mutate(År = ifelse(n == 2, str_c(År, letter), År)) %>%
select(År, namn, value) %>%
pivot_wider(names_from = namn)
}To plot times/dates in ggplot with lubridate, the period needs to be
converted to seconds using for example the duration function. The to set
the scales, use scale_x_time/scale_y_time. To customize breaks and
labels, use the scales library. In this case we use
breaks = scales::breaks_width("15 min"), labels =scales::label_time(format = "%H:%M").
tibble(minutes = 100:290,
time_parsed = seconds_to_period(dminutes(minutes)) %>% as.numeric() %>% duration) %>%
mutate(pace = get_pace("marathon", minutes) %>% as.numeric()%>% duration) %>%
ggplot(aes(x= time_parsed, y = pace)) +
geom_line() +
scale_x_time(breaks = scales::breaks_width("15 min"), labels =scales::label_time(format = "%H:%M")) +
scale_y_time(breaks = scales::breaks_width("0.25 min"), labels =scales::label_time(format = "%M:%S"))library(ggpubr)
ggarrange(p1, p2, p3)
# with common legend:
ggarrange(p1, p2, p3, ncol=3, common.legend = T, legend="right")library(tidyverse)
library(ggpubr)
# with common legend
p11 <- iris %>% ggplot(aes(Sepal.Length, Sepal.Width, color=Species)) + geom_point()
# with common legend
p1 <- ggarrange(p11 ,
p11 ,
p11 ,
ncol=1, nrow=3, legend = "none"
)
p2 <- ggarrange( p11 , legend = "none")
p3 <- ggarrange(
p11,
p11 ,
p11 ,
ncol=1,nrow=3, common.legend = T, legend="right"
)
ggarrange(p1, p2, p3, ncol=3, common.legend = T, legend="right")Generates:
library(tidyverse)
p11 <- iris %>% ggplot(aes(Sepal.Length, Sepal.Width, color=Species)) + geom_point()
# change title in legend:
p11 + labs(color = "Hej") # or fill/group
# Alternatively:
p11 + scale_color_discrete("Hej")
p11 + scale_fill_discrete("Hej") # if fill=Species
p11 + scale_color_manual("Hej") + scale_color_discrete(labels = c("A", "B", "C")) # for ggplot(aes(color=category))
+ scale_fill_discrete(labels = c("A", "B", "C")) # for ggplot(aes(fill=category))library(tidyverse)
library(RColorBrewer)
p11 <- iris %>% ggplot(aes(Sepal.Length, Sepal.Width, color=Species)) + geom_point()
p11_c <- ggplot(iris, aes(x=Sepal.Length, y=Petal.Length, color=Sepal.Width)) + geom_point()
# choose custom color palette
p11 + scale_color_brewer("Hej",palette = "Reds")
# continous:
p11_c + scale_color_gradient(low = "white", high = "red")
# alternatively:
num_categories <- 3
cls_size <- brewer.pal(num_categories, "Reds")
p11 + scale_color_manual("Hej", values = cls_size, drop=FALSE)
# or if fill=Species, use:
# scale_fill_manual("Hej", values = cls_size, drop=FALSE)
# scale_linetype_manual()
# choose a pre-defined color palette:
p11 + scale_color_viridis_d("Hej")
p11 + scale_color_grey()# stat=identity: use own y-values, rather than counting the aggregate number of rows for each x (stat=count)
# position=dodge : place bars side by side instead of stacked
geom_bar(stat="identity", position = "dodge")
# add labels above bar plot, for example percentage value
+ geom_text(aes(label= str_glue("{value*100}%") ),
position=position_dodge(width=0.9),
vjust=-0.25) Dodging overlapping labels:
p <- ggplot(mpg) +
geom_bar(aes(x = manufacturer)) +
theme(axis.text.x = element_text(size = 11))
# Use guide_axis to dodge the labels
p +
scale_x_discrete(guide = guide_axis(n.dodge = 2))Introduced in ggplot2 3.3.0. A binning scale takes continuous data and makes it discrete by assigning each data point to a bin. Allows continuous data to be used with discrete palettes
p <- ggplot(mpg) +
geom_point(aes(displ, cty, size = hwy, colour = hwy))
p +
scale_size_binned()
# show outermost ticks:
p +
scale_size_binned(guide = guide_bins(show.limits = TRUE))
p +
scale_colour_binned()
# place data at the center of the bin they belong to:
p +
scale_x_binned()
# this facilitates to create histograms with tick-marks between the bars:
ggplot(mpg) +
geom_bar(aes(displ)) +
scale_x_binned()# procedure is a text column
f <- function(x) factor(x, levels = unique(x))
reshape2::melt(tmp, id.vars = "Procedure") %>%
ggplot(aes(x=f(Procedure), y=value*100, fill=variable)) library(tidyverse)
library(sf)
library(RColorBrewer)
# Data för Sverigekarta
tmp1 <- tempfile()
download.file("http://api.thenmap.net/v2/se-7/geo/2020-08-14", destfile = tmp1)
county <- read_sf(tmp1)
county <- st_transform(x = county, crs = 3006)
county <- merge(county, antal, by.x = "id", by.y = "Kommun", all.x=T)
county$size <- cut(county$Antal, breaks = c(0, 10, 100, 1000, Inf),
labels = c("1-10", "11-100", "101-1000", ">1000"))
cls_size <- brewer.pal(4, "Reds")
ggplot() +
geom_sf(data = county, aes(fill = size)) +
theme_void() +
scale_fill_manual("Municipality size", values = cls_size) | Distribution | cdf |
Inverse cdf/quantile |
pdf |
Generate a random variable |
|---|---|---|---|---|
| Beta | pbeta |
qbeta |
dbeta |
rbeta |
| Binomial | pbinom |
qbinom |
dbinom |
rbinom |
| Chi-Square | pchisq |
qchisq |
dchisq |
rchisq |
| Discrete Uniform | extraDistr::pdunif |
extraDistr::qdunif |
extraDistr::ddunif |
extraDistr::rdunif |
| Exponential | pexp |
qexp |
dexp |
rexp |
| F | pf |
qf |
df |
rf |
| Gamma | pgamma |
qgamma |
dgamma |
rgamma |
| Geometric | pgeom |
qgeom |
dgeom |
rgeom |
| Logistic | plogis |
qlogis |
dlogis |
rlogis |
| Log Normal | plnorm |
plnorm |
dlnorm |
rlnorm |
| Negative Binomial | pnbinom |
qnbinom |
dnbinom |
rnbinom |
| Normal | pnorm |
qnorm |
dnorm |
rnorm |
| Poisson | ppois |
qpois |
dpois |
rpois |
| Student t | pt |
qt |
dt |
rt |
| Studentized Range | ptukey |
qtukey |
- | - |
| Uniform | punif |
qunif |
dunif |
runif |
| Weibull | pweibull |
qweibull |
dweibull |
rweibull |
| Wilcoxon Rank Sum Statistic | pwilcox |
qwilcox |
dwilcox |
rwilcox |
| Wilcoxon Signed Rank Statistic | psignrank |
qsignrank |
dsignrank |
rsignrank |
For more distributions not included in base R, see the package
extraDistr .
library(tidyverse)
library(patchwork)
options(scipen = 999)
mu <- 50
sd <- 5
n <- 1000
set.seed(10)
df <- tibble(
x = seq(1, 100, length.out = n),
p = seq(0, 1, length.out = n),
pdf = dnorm(x, mean = mu, sd = sd), # f(x) = P(X = x)
cdf = pnorm(x, mean = mu, sd = sd), # F(x) = P(X <= x)
q = qnorm(p, mean = mu, sd = sd), # F^{-1}(p) = x
X = rnorm(n, mean = mu, sd = sd) # generate a random variable
)
p_pdf <- df %>%
ggplot(aes(x = x, y = pdf)) +
geom_point(size = 0.5) +
theme_light() +
labs(
title = "The pdf (dnorm): P(X = x)",
subtitle = str_glue("n = {nrow(df)}, mu = {mu}, sd = {sd}")
)
p_cdf <- df %>%
ggplot(aes(x = x, y = cdf)) +
geom_point(size = 0.5) +
theme_light() +
labs(
title = "The cdf (pnorm): F(x) = P(X <= x)",
subtitle = str_glue("n = {nrow(df)}, mu = {mu}, sd = {sd}")
)
p_q <- df %>%
ggplot(aes(x = x, y = q)) +
geom_point(size = 0.5) +
theme_light() +
labs(
title = "The quantile / inverse cdf (qnorm)",
subtitle = str_glue("n = {nrow(df)}, mu = {mu}, sd = {sd}")
)
p_X <- df %>%
ggplot(aes(x = X)) +
geom_density() +
theme_light() +
labs(
title = "Sample generated from X ~ N(50, 5)",
subtitle = str_glue("n = {nrow(df)}, mu = {mu}, sd = {sd}")
)
(p_cdf + p_q )/
( p_pdf + p_X)The cdf and the inverse cdf are related by
I.e. given a number (probability) between 0 and 1, return the p-th quantile, i.e. the x-value on the plot. Consider also the difference between the ‘theoretical’ quantile of the population (q) compared to the observed sample quantile (qs).
options(scipen = 999)
plotnn <- function(n, mu, sd) {
set.seed(10)
X <- rnorm(n, mu, sd) # Generate random variable
p <- 0.5 # Let the probability be 50% (the 50th percentile)
q <- qnorm(p, mu, sd) # inverse cdf, theoretical quantile
cdf <- pnorm(q, mu, sd) # cdf should be equal to p
xbar <- mean(X) # sample mean
xmedian <- median(X) # sample median
sd(X)
q_sample <- qnorm(p, xbar, sd(X)) %>% round(3)
0.5 == pnorm(q_sample, xbar, sd(X)) # TRUE
plot1 <- X %>%
enframe() %>%
ggplot(aes(x = value)) +
geom_density(fill = "gray") +
theme_minimal()+
theme(legend.position="top")
# extract x,y values from plot in order to shade the area
d <- ggplot_build(plot1)$data[[1]]
d <- d %>% mutate(color = case_when(
x <= q & x > q_sample~ "P(X <= q)",
x <= q_sample ~ "P(X <= qs)"
))
plot1 +
geom_area(data = subset(d, x <= q), aes(
x = x, y = y,
fill = factor(color)
)) +
annotate("text", x = q + 1, y = 0, label = str_glue("q = {q}")) +
annotate("text", x = q - 1, y = 0 + 0.005, label = str_glue("q_sample = {q_sample}")) +
geom_vline(xintercept = q, linetype = "dashed") +
geom_vline(xintercept = q_sample, linetype = "dotted") +
scale_color_discrete("cdf:") +
labs(
title = "Relationship between cdf and inverse cdf (the quantile q)",
subtitle = str_glue("X ~ N({mu}, {sd}), n = {n}. xbar = {round(xbar,4)}, median = {round(xmedian,4)}"),
x = "x",
fill = "cdf"
) +
scale_fill_manual(breaks = c("P(X <= q)", "P(X <= qs)"),
values = c("#FC4E07","#00AFBB"))
}
mu <- 50
sd <- 5
plotnn(10, mu, sd)plotnn(100, mu, sd)plotnn(1000, mu, sd)plotnn(100000, mu, sd)Here is a QQ-plot to illustrate the observed quantiles (y-axis) vs the
theoretical quantiles (x-axis). This can be used to assess normality.
Note that the plot looks the same even if the values are z-transformed
or not (verify it yourself if you like by changing
ggplot(aes(x = qq, y = x)) to ggplot(aes(x = qq, y = x_scaled))).
mu <- 50
sd <- 5
plotqq <- function(n, mu, sd) {
set.seed(10)
X <- rnorm(n, mu, sd)
d <- X %>%
enframe(name = NULL, value = "x") %>%
mutate(across(x, scale, .names = "{.col}_scaled")) %>% # z-transform to mean zero and unit variance
arrange(x) %>%
mutate(j = 1:length(X),
prob_level = (j-0.5)/length(X), # 0.5 is the 'continuity' correction
qq = qnorm(prob_level) # standard normal quantile,
)
d %>%
ggplot(aes(x = qq, y = x)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x, se = FALSE) +
labs(y = "Observed quantiles",
x = "Theoretical quantiles",
subtitle = str_glue("n = {n}, mu = {mu}, sd = {sd}")) +
theme_light()
}
p1 <- plotqq(10,mu,sd)
p2 <- plotqq(100,mu,sd)
p3 <- plotqq(1000,mu,sd)
p4 <- plotqq(100000,mu,sd)
(p1+p2) /
(p3+p4)The table below shows a set of possible values (domains) for the quantile(x) and percentile(x) functions, while the quartile is always between 0-4.
| Quartile | Quantile | Percentile |
|---|---|---|
| 0 | 0 | 0 |
| 1 | 0.25 | 25 |
| 2 | 0.5 | 50 (median) |
| 3 | 0.75 | 75 |
| 4 | 1 | 100 |
This does not say that a quantile varies between 0 and 1, and percentile between 0 and 100. Quantiles can go from anything to anything. Link
Also note that for samples from the normal distribution, the median may not equal the sample quantile but they should not be far apart.
The sample variance can be expressed in many ways, here are the two most common ways:
Proof:
Where
The sum of the deviations from the mean of a measurement is always equal to 0
–
Conditional probability of event A, given that event B has occurred:
$$
\begin{align}
P(A|B) = \frac{P(A \cap B)}{P(B)}
\end{align}
$$ Multiplicative law of probability:
If A and B are independent, it holds that
Additive law of probability:
The collection of sets
$$ \begin{align} P(A) = \sum_{i=1}^k P(A|B_i)P(B_i) \end{align} $$ ### Bayes Rule:
Assume that
–
A random variable, denoted in capitals, is a real-valued function for
which the domain is a sample space. For example, let
–
Survey sampling is a statistical process (statistisk undersökning) characterized by the following:
-
A well-defined finite set of elements, a population. It must be clear which elements are a part of the population you wish to study.
-
The elements are measurable and there is a need for information about some population parameters.
-
A sampling frame (urvalsram) is needed, i.e. an operational representation of the elements. For example a hierarchy of sampling units that associate each element with a sampling unit.
-
A sample (stickprov) of elements is done by sampling sampling units from the sampling frame accoridning to some sample design (urvalsdesign), such that each element has a known positive probability of being selected. An exception being a total survey (totalundersökning) where all elements are selected.
-
The sampled elements are observed (measured) with regards to the set of variables that are of interest for the survey accordning to a measurement plan.
-
The collected observations are finally used to estimate population parameters (total, mean, percent, quotients, regression coefficients, …) where information is needed.
Typically, the set notation is used in survey sampling. The variable
(undersökningsvariabeln) is denoted as
The population is
Let
The parameter (målstorhet)
The mean is a function of the total:
To estimate the parameters, a sample
Without replacement, ie an object can only be included in a sample once.
The definition of OSU is that each possible combination of samples drawn
of (fixed) size
For example, consider the objects A, B, C, D, E (
# A tibble: 10 × 7
Nr A B C D E `p(s)`
<int> <chr> <chr> <chr> <chr> <chr> <dbl>
1 1 "A" "B" "C" "" "" 0.1
2 2 "A" "B" "" "D" "" 0.1
3 3 "A" "B" "" "" "E" 0.1
4 4 "A" "" "C" "D" "" 0.1
5 5 "A" "" "C" "" "E" 0.1
6 6 "A" "" "" "D" "E" 0.1
7 7 "" "B" "C" "D" "" 0.1
8 8 "" "B" "C" "" "E" 0.1
9 9 "" "B" "" "D" "E" 0.1
10 10 "" "" "C" "D" "E" 0.1
Note for example that the probability of selecting A is
Inklusionssannolikhet (första ordningens).
The probability of selecting element
In the example above for
This can be shown by asking: what is the probability of an event happening k times in n trials, without replacement? We can use the hypergeometric distribution.
An indicator variable can be used to indicate if element k is included
in the sample:
Also note that:
since
The variance of the inclusion probability is
–
The probability of selecting both element
which is
For example, from the example table we can visually see that selecting
both A and B has a probability of 0.3 (
# A tibble: 10 × 7
Nr A B C D E `p(s)`
<int> <chr> <chr> <chr> <chr> <chr> <dbl>
1 1 "A" "B" "C" "" "" 0.1
2 2 "A" "B" "" "D" "" 0.1
3 3 "A" "B" "" "" "E" 0.1
4 4 "A" "" "C" "D" "" 0.1
5 5 "A" "" "C" "" "E" 0.1
6 6 "A" "" "" "D" "E" 0.1
7 7 "" "B" "C" "D" "" 0.1
8 8 "" "B" "C" "" "E" 0.1
9 9 "" "B" "" "D" "E" 0.1
10 10 "" "" "C" "D" "E" 0.1
which is shown to be equal to
The HT-estimator is a general unbiased estimator regardless of the sampling design.
where
The design weight can be interpreted as how many objects a certain
object from the sample shall represent. If
For example, for a sample of 100 schools from a population of 1000 schools, each school in the sample represents 10 schools in the population.
Note that:
-
Indcator variable
$I _k$ was added so sum over s changed to sum over U -
$y$ is regarded as a fixed constant in this type of survey sampling. The randomness is instead introduced from the sampling design. The advantage is that no assumptions are needed (in the case of full response).
Note that the mean is estimated with
In OSU,
Variance of a linear combination of variables general formula:
Where the variance of the inclusion indicator variable is
and the covariance
Also note that
Collect expressions
Notice that the two sums could be simplified to a single sum because
when
It can be shown that
where
Proof:
Where
and
Simplify expression outside the parenthesis
Note that the right term inside the parenthesis can be simplified as
follows:
It can now be simplified as following:
An algorithm for performing sampling (urvalsdragning). The goal of the algorithm is to fulfill the sampling design.
-
Dragsekventiella: En serie slumpmässiga försök från hela populationen. Ex Bingolotto med tombola med numren 1 till 37.
-
Listsekventiella: Urvalsramens enheter gås igenom enhet för enhet. Ett slumpmässigt försök utförs för varje enhet för att avgöra om enheten ska ingå i urvalet eller inte. (most common)
For OSU, the most common algorithm is Sunter (1977). The algorithm simply assigns a random number drawn from uniform distribution (0,1) as a key to each item, then sorts all items using the key and selects the smallest k items.
Here is an implementation in R/tidyverse:
library(tidyverse)
library(survey)
data(api)
k <- 3
# if you have stratum, you can also create a column 'small n'/'nh' with the number of observation to select from each stratum and use that instead of k
selection <- apistrat %>%
mutate(unif = runif(n = n())) %>%
# sorterar data on stratum (stype) and uniform random variable value
arrange(stype, unif) %>%
group_by(stype) %>%
mutate(x = sequence(n())) %>%
# vi väljer ut nh första raderna per stratum
filter(x <= k) %>%
ungroup()
selection %>% select(1:4, enroll, unif, x)# A tibble: 9 × 7
cds stype name sname enroll unif x
<chr> <fct> <chr> <chr> <int> <dbl> <int>
1 19647256015473 E Lowell Elementa Lowell Elementary 450 3.54e-4 1
2 19647336018568 E One Hundred Twe One Hundred Twelfth… 446 6.31e-4 2
3 01612596001754 E Crocker Highlan Crocker Highlands E… 266 1.32e-3 3
4 34674473435930 H Mira Loma High Mira Loma High 1148 7.58e-2 1
5 34674393437555 H Sacramento High Sacramento High 1389 1.21e-1 2
6 31669513134657 H Lincoln High (C Lincoln High (Char) 599 1.22e-1 3
7 38684786062038 M Everett Middle Everett Middle 491 7.31e-2 1
8 19648406021125 M Benton (Reginal Benton (Reginald M.… 775 7.53e-2 2
9 01611926066476 M King (Martin Lu King (Martin Luther… 618 8.28e-2 3
Elements in the frame (ramen)
Let
Schema: element
The sample size is random. The expected sample size it
Systematiskt urval. This design is not measurable because the variance cannot be estimated.
Let 𝑎 be sampling interval
Let 𝑛 be the integer part by dividing 𝑁/𝑎 and let 𝑐 be the rest
Then, 𝑁=𝑛𝑎+𝑐
-
200=20×10+0
-
201=20×10+1
-
207=20×10+7
If rest 𝑐>0 the sample will be 𝑛 or 𝑛+1
In 3) above, 𝑛=21 with random start between 1 and 7 In 3) above, 𝑛=20 with random start between 8 and 10
This sampling design can be useful for sampling in a geographical area, for example a grid of 1x1 meter squares are the objects. Compared to OSU the sampling may be more evenly distributed, whereas OSU can sample many observations in a certain location.
If
A list sequential schema is given by:
Let
-
pps (with replacement)
-
$\pi$ps (without replacement)
library(devtools)
devtools::install_github(repo = "DiegoZardetto/ReGenesees")
library(ReGenesees)
# skapa designinformation
design4 <- e.svydesign(data = as.data.frame(data),
ids = ~id,
strata = ~stratum,
weights = ~designvikt,
fpc = ~StoraN
)
design4
#utb 3535715 2654451 1488358
# skapa en population template, det behövs för kalibrering
aux4 <- pop.template(design4, calmodel=~(aldkon+utb-1), partition = F)
aux4
aux4[1] <- 841269
aux4[2] <- 950487
aux4[3] <- 1236159
aux4[4] <- 830221
aux4[5] <- 796725
aux4[6] <- 911748
aux4[7] <- 1211723
aux4[8] <- 900192
aux4[9] <- 2654451 # OBS! utb kategori 2
aux4[10] <- 1488358 # OBS! utb kategori 3
aux4
# kalibrering utförs här
descal4 <- e.calibrate(design = design4,
df.population = aux4,
calmodel= ~(aldkon+utb-1),
partition = FALSE,
calfun = "linear",
bounds = c(-100, 100),
aggregate.stage = NULL
)
summary(descal1b)
weights(descal1b) %>% unique
data$ald
# Estimationsanrop
svystatTM(descal4, ~y, by = ~ald)
svystatTM(descal4, ~y, by = ~ald, estimator = c("Mean"))# delta: difference in means
# Number of subjects needed to obtain 80% power
power.t.test(delta = 10, sd = 19, power = 0.8, sig.level = 0.05)
# calculate power from sample size:
power.t.test(delta = 10, sd = 19, n=100, sig.level = 0.05)Also known as the Mann-Whitney U test.
wilcox.test(df$num ~ temp$group)Tests difference in means (two-sample test). Assumptions: your data values are independent, are randomly sampled from two normal populations. The two independent groups are by default treated as not having equal variances in the t.test function but can be changed.
library(dplyr)
# sleep data:
# Data which show the effect of two soporific drugs
# (increase in hours of sleep compared to control) on 10 patients.
# extra = numeric increase in hours of sleep
# group = drug given
sleep %>% group_by(group) %>% dplyr::slice_sample(n = 3)# A tibble: 6 × 3
# Groups: group [2]
extra group ID
<dbl> <fct> <fct>
1 -0.1 1 5
2 -0.2 1 3
3 3.7 1 7
4 -0.1 2 5
5 4.6 2 9
6 3.4 2 10
t.test(extra ~ group, data = sleep) Welch Two Sample t-test
data: extra by group
t = -1.8608, df = 17.776, p-value = 0.07939
alternative hypothesis: true difference in means between group 1 and group 2 is not equal to 0
95 percent confidence interval:
-3.3654832 0.2054832
sample estimates:
mean in group 1 mean in group 2
0.75 2.33
In this case,
cor.test()For testing the null hypothesis of independence of rows and columns in a contingency table with fixed marginals.
fisher.test( matrix(c(2,10,20,3),nrow=2,ncol=2) )
# odds ratio: 0.035, p=0.00008124In Python:
import scipy.stats as stats
oddsratio, pvalue = stats.fisher_exact([[2, 10], [20, 3]])
# (0.03, 8.124495626949326e-05)Kappa measures the agreement between two variables, taking into account the agreement that would happen by chance.
library(psych)
tab<-matrix(c(12,20,20,12),ncol=2,nrow=2)
# [,1] [,2]
#[1,] 12 20
#[2,] 20 12
psych::cohen.kappa(tab) # -0.25binom.test(40, 95) # num successes, num trialsWith CI in parenthesis for.ex. (10.1-15.5%) calculated from percentage of success:
library(stringr)
calc_conf <- function(perc, n){
test <- round(perc*n)
res <- binom.test(test, n)
lower <- res$conf.int[1] %>% round(3) *100
upper <- res$conf.int[2] %>% round(3) *100
lower <- format(lower, nsmall = 1)
upper <- format(upper, nsmall = 1)
return(str_glue("({lower}-{upper}%)"))
}
calc_conf(0.42, 95) # (32.0-52.7%)library(DescTools)
# x = number of successes in group 1 or 2, n = number of trials in group 1 or 2
BinomDiffCI(x1=40,n1=95,x2=2,n2=47)Nicely formatted CI calculated from percentage of success:
library(stringr)
bindiff <- function(perc_1, n_1, perc_2, n_2){
test1 <- round(perc_1*n_1)
test2 <- round(perc_2*n_2)
res <- BinomDiffCI(test1,n_1,test2,n_2)
val <- res[1] %>% round(3) *100
lower <- res[2] %>% round(3) *100
upper <- res[3] %>% round(3) *100
val <- format(val, nsmall = 1)
lower <- format(lower, nsmall = 1)
upper <- format(upper, nsmall = 1)
return(
list(test1=test1,test2=test2,
ci = str_glue("{val}% ({lower}-{upper}%)")
))
}
bindiff(0.42,95,0.042,47) # 37.8% (24.2-48.0%)Proportional odds: the effect of the independent variables on the dependent variable is constant for all levels in the dependent variable.
https://m-clark.github.io/mixed-models-with-R/random_intercepts.html
Random intercepts model: group-specific intercepts with own unique effects. E.g. (1 | student) the intercept 1 is allowed to vary between student to student. The random effect is to the left of |.
library(survival) # survival curves
fit1 <- survfit(Surv( time , survival ) ~ variable, data=df)
# Survival plot:
#install.packages("survminer")
library(survminer)
ggsurvplot(fit1)library(survival)
fit1 <- coxph(Surv( time , survival ) ~ variable, data=df)
summary(fit1)library(tidyverse)
is_outlier <- function(x) {
return(x < quantile(x, 0.25,na.rm = T) - 1.5 * IQR(x, na.rm = T) | x > quantile(x, 0.75, na.rm = T) + 1.5 * IQR(x,na.rm=T))
}
# example: number of outlier per variable
mtcars %>%
summarise(across(where(is.numeric), is_outlier)) %>%
summarise(across(everything(), ~sum(.==TRUE))) %>%
t() [,1]
mpg 1
cyl 0
disp 0
hp 1
drat 0
wt 3
qsec 1
vs 0
am 0
gear 0
carb 1
Note: outliers in boxplot() is computed differently. In that case, use
boxplot.stats()$out to see outlier values.
Never use for (i in 1:lenght(n)) because it will fail if length(n)
is 0, i.e. it will evaluate to 1 0.
Use seq_along or seq_len instead.
For faster looping.
out <- vector("list", length(n))
# out <- vector("numeric", length(n)) #alternatively
for (i in seq_len(n)) {
out[[i]] <- 1 + i
}Using deparse + substitute
print_path <- function(path) {
print(stringr::str_glue("{deparse(substitute(path))} = {path}"))
}Code - a sequence of symbols/constants/calls that will return a result if evaluated. Code can be:
(i) Evaluated immediately (Standard Eval), (ii) Quoted to use later (Non-Standard Eval).
Quosure- An expression that has been saved with an environment (aka a
closure).
A quosure can be evaluated later in the stored environment to return a
predictable result.
a <- 1
b <- 2
q <- rlang::quo(a+b)
# <quosure>
#expr: ^a + b
#env: globalEvaluate it with rlang::eval_tidy(q), returning 3.
Useful when doing ggplot functions. For example with this function you
can create a function like this f(mtcars, disp, mpg ), where disp and
mpg are columns within the dataframe mtcars (included in tidyverse). !!
uncoutes the symbol.
library(tidyverse)
f <- function(df,a,b){
a <- rlang::enquo(a)
b <- rlang::enquo(b)
# !! unquotes the symbol
df %>%
ggplot(aes(x=!!a,y=!!b)) +
geom_point()
}
mtcars
f(mtcars, disp, mpg )Returns a tibble of useful information from Swedish Police reports given location, date or event. More info here about what you can do: https://polisen.se/om-polisen/om-webbplatsen/oppna-data/api-over-polisens-handelser/
library(httr)
library(tidyverse)
library(lubridate)
library(jsonlite)
get_polisen <- function(.date = NULL, .location=NULL, .events = NULL, .preprocess = T) {
base_url <- "https://polisen.se/api/events?"
url <- base_url
if (!is.null(.events)) {
events <- str_c(.events, collapse = ";")
url <- str_c(base_url, "type=", events)
}
if (!is.null(.date)) {
url <- str_c(url, "&DateTime=", .date)
}
if (!is.null(.location)) {
locations <- str_c(.location, collapse = ";")
url <- str_c(url, "&locationname=", locations)
}
resp <- httr::GET(as.character(url))
if (httr::http_status(resp)$category != "Success") {
print(resp)
stop("Http status not success")
}
df <- resp %>%
content(as = "text") %>%
jsonlite::fromJSON() %>%
as_tibble()
if (.preprocess & nrow(df) != 0) {
df <- df %>%
mutate(datetime = lubridate::as_datetime(datetime, tz = "Europe/Stockholm"),
url = str_glue("https://polisen.se{url}"),
summary2 = str_glue("{summary}\n<a href=\"{url}\">Mer info</a>")
)
# separate coordinates into two columns, rename duplicate column name
df$location <- df$location %>%
as_tibble() %>%
separate(gps, c("lat", "lng"), sep = ",", convert = T) %>%
dplyr::rename(location_name = name) %>%
# add some noise to coordinates, since all coordinates for a given location are the same.
# otherwise the points will overlap on the map.
# the coordinates only show the coordinate of an area, never an exact location
mutate(lat = jitter(lat), lng = jitter(lng) )
df <- df %>%
unnest(location)
}
return(df)
}
# get all events:
get_polisen()
# Filter by a vector of events
get_polisen(.date = "2022-06", .location = "Varberg", .events = c("Rån", "Trafikolycka"))
# filter by a vector of locations
get_polisen(.date = "2022-04", .location = c("Varberg","Stockholm"))
# filter by year, year-month or year-month-day:
get_polisen(.date = "2022-06-05", .location = "Linköping")
# # A tibble: 1 x 9
# id datetime name summary url type location_name lat lng
# <int> <dttm> <chr> <chr> <glue> <chr> <chr> <dbl> <dbl>
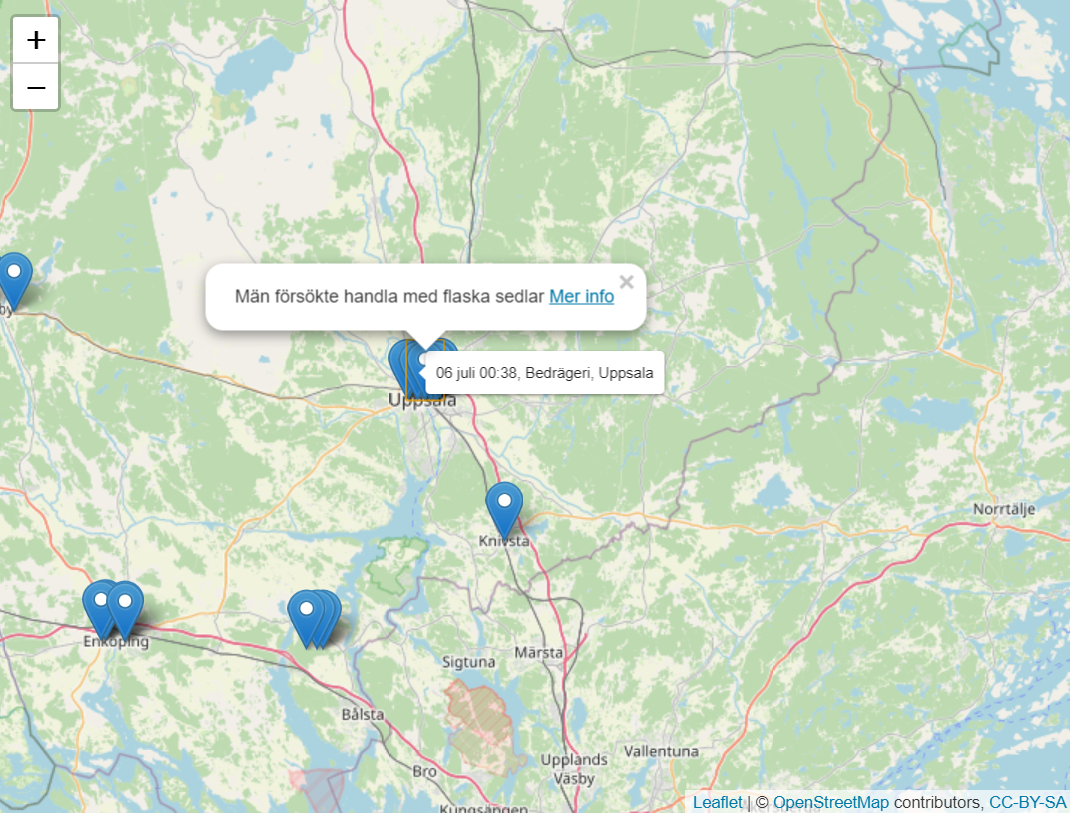
# 342083 2022-06-05 11:31:12 05 juni 11:31, Rattfylleri, Linköping Trafikant uppmä~ https://polisen.se~ Rattf~ Linköping 58.4 15.6The following code will plot an interactive map of all the coordinates retrieved via the Polisen API function, with popups containing information and links for more information.
Note that the coordinates from Polisen only show the coordinates of a
city or area, not an exact location. Within get_polisen(), numeric
noise is added to the coordinates in order to be able show all
coordinates on a plot (otherwise they will all overlap for a given
area). In order to find out the exact location or area within a city,
you would have to click on ‘Mer info’ where it will probably be given in
more detail in the police report.
# install.packages("leaflet")
library(leaflet)
library(tidyverse)
get_polisen(.date = "2022-06", .location = "Uppsala") %>%
leaflet() %>%
addTiles() %>%
addMarkers(lng = ~lng, lat = ~lat, popup = ~summary2, label = ~name)See code for more details. Given a coordinate, retrieve traffic information within a 10 000 meter radius and display on an interactive map. Requires that you register for an api-key at Trafikverket. For more info, see here.
x <- get_traffic_info(.x = "6398983", .y = "320011")
x %>%
plot_traffic()unlink("path/*", recursive = TRUE)
unlink("path/*") # deletes files only, no directories# set pattern = ".*out.*" to target specific directories named out
out_dirs <- list.files(str_glue("C:/path1/path2"), full.names = T, recursive = T, pattern = ".*out.*", include.dirs = T)
out_dirs <- str_c(out_dirs, "/*")
# out_dirs contains: "C:/path1/path2/ax/out/*" "C:/path1/path2/dk/out/*" "C:/path1/path2/EA/out/*" "C:/path1/path2/EU/out/*" ...
unlink(out_dirs, recursive = T) # removes files and dirs under "C:/path1/path2/{ax,dk,EA,EU,...}/out/*"basename("C:/px/hej.txt")
# hej.txt
dirname("C:/px/hej.txt")
# "C:/px"If you’re a windows user, this is good to know if you need to do any form of automation on system files.
dir C:\Users\emiwes\Documents\experiments -Recurse | Select-String -pattern "my search"This example removes all files containing _flags in filename:
Dir | Rename-Item –NewName { $_.name –replace “_flags“,”” }system("powershell ls -rec H:\\Documents\\")Add ls -rec for subfolders and for ex. -Include *.txt for including txt files only
system("powershell (ls H:\\Documents\\multiple_tests\\*) |
Foreach-Object{ $f=$_; (gc $f.PSPath) |
Foreach-Object {$_ -replace '\\\"hej', 'hej' } |
Set-Content $f.PSPath
}
")Run this in the folder you want to make changes in
foreach($i in ls -recurse -filter "*.R") {
$temp = Get-Content $i.fullname
Out-File -filepath $i.fullname -inputobject $temp -encoding utf8 -force
}Git is a distributed version control system that is really useful to know.
Note: [Using git version 2.30.0.windows.2]
# open global .gitconfig in ~/.gitconfig. To find where ~ is on windows, type echo $HOME
git config --global --edit
# create and switch to branch:
git checkout -b main/emil_westin
# push to your current branch:
git push origin HEAD
# push to main directly:
git push origin HEAD:main
# show changes:
git diff
# add everything to staged:
git add -A
# unstage all files:
git reset
# unstage specific file:
git restore --staged file.txt
# commit:
git commit -m "message"
# to modify previous commit, for ex include a new file, type:
git commit --amend
# if you already have a folder that you want to track with version conrol and associate with a remote repository:
git remote add origin https://linktoyourremotereporitory
# clone a repository (get a local copy incl all version history):
git clone https://linktoremotereporitory
Nice extra things to add in the ~/.gitconfig to add color in the terminal:
[color]
ui = auto
[color "branch"]
current = yellow reverse
local = yellow
remote = green
[color "diff"]
meta = yellow bold
frag = magenta bold
old = red bold
new = green bold
[color "status"]
added = yellow
changed = green
untracked = cyan
[0-9]+\.?[0-9]{0,}
Pros of using lubridate: more informative output, no need to re-invent
the wheel.
Pros of doing manual calculation: more control/understanding how it is calculated.
library(tidyverse)
library(lubridate)
get_pace <- function(distance, target_duration, decimals = 1, style = "below") {
if (distance == "marathon") dist <- 42.195 # km
if (distance == "half marathon") dist <- 21.0975
if (str_detect(distance, "[0-9.]+")) dist <- as.numeric(distance)
s <- target_duration*60 # convert to seconds
if (style == "below") pace <- (s-1)/dist
if (style == "exact") pace <- s/dist
res <- seconds_to_period(pace)
return(res %>% round(decimals))
}
# pace to complete marathon under 4h 30min (4h 29min 59 seconds)
get_pace("marathon", 270)
# "6M 24S"
# works with kilometers as well
get_pace(42.195, 270)
# "6M 24S"
# and miles
get_pace(26.2, 270)
# "10M 18S"
# supply number of decimals for more details
get_pace("marathon", 270, decimals = 2)
# "6M 23.91S"
# pace to complete marathon in exactly 4h 30min
get_pace("marathon", 270, 2, "exact")
# "6M 23.93S"
# generate a table
library(kableExtra)
options(knitr.kable.NA = '')
tibble(minutes =
c(
seq(from = 13, to = 25, by = 1),
seq(from = 27.5, to = 30, by = 2.5),
seq(from = 35, to = 60, by = 5),
seq(from = 65, to = 115, by = 5),
seq(from = 120, to = 360, by = 15)
)
,
parsed_time = seconds_to_period(duration(minutes, "minutes")-1) %>% as.character() %>% str_remove(" 0S"),
marathon = get_pace("marathon", minutes) %>% as.character(),
halfmarathon = get_pace("half marathon", minutes) %>% as.character(),
`10k` = get_pace(10, minutes) %>% as.character(),
`5k` = get_pace(5, minutes) %>% as.character()
) %>%
mutate(marathon = ifelse(between(minutes, 120, 360), marathon, NA_character_)) %>%
mutate(halfmarathon = ifelse(between(minutes, 60, 180), halfmarathon, NA_character_)) %>%
mutate(`10k` = ifelse(between(minutes, 25, 80), `10k`, NA_character_)) %>%
mutate(`5k` = ifelse(between(minutes, 10, 40), `5k`, NA_character_)) %>%
select(-minutes) %>%
kableExtra::kable(format = "pipe")The following is a table of target times for kilometer paces (one second below the given target time).
| target time | marathon | halfmarathon | 10k | 5k |
|---|---|---|---|---|
| 13M | 2M 35.8S | |||
| 14M | 2M 47.8S | |||
| 15M | 2M 59.8S | |||
| 16M | 3M 11.8S | |||
| 17M | 3M 23.8S | |||
| 18M | 3M 35.8S | |||
| 19M | 3M 47.8S | |||
| 20M | 3M 59.8S | |||
| 21M | 4M 11.8S | |||
| 22M | 4M 23.8S | |||
| 23M | 4M 35.8S | |||
| 24M | 4M 47.8S | |||
| 25M | 2M 29.9S | 4M 59.8S | ||
| 27M 30S | 2M 44.9S | 5M 29.8S | ||
| 30M | 2M 59.9S | 5M 59.8S | ||
| 35M | 3M 29.9S | 6M 59.8S | ||
| 40M | 3M 59.9S | 7M 59.8S | ||
| 45M | 4M 29.9S | |||
| 50M | 4M 59.9S | |||
| 55M | 5M 29.9S | |||
| 1H 0M | 2M 50.6S | 5M 59.9S | ||
| 1H 5M | 3M 4.8S | 6M 29.9S | ||
| 1H 10M | 3M 19S | 6M 59.9S | ||
| 1H 15M | 3M 33.2S | 7M 29.9S | ||
| 1H 20M | 3M 47.5S | 7M 59.9S | ||
| 1H 25M | 4M 1.7S | |||
| 1H 30M | 4M 15.9S | |||
| 1H 35M | 4M 30.1S | |||
| 1H 40M | 4M 44.3S | |||
| 1H 45M | 4M 58.6S | |||
| 1H 50M | 5M 12.8S | |||
| 1H 55M | 5M 27S | |||
| 2H 0M | 2M 50.6S | 5M 41.2S | ||
| 2H 15M | 3M 11.9S | 6M 23.9S | ||
| 2H 30M | 3M 33.3S | 7M 6.5S | ||
| 2H 45M | 3M 54.6S | 7M 49.2S | ||
| 3H 0M | 4M 15.9S | 8M 31.9S | ||
| 3H 15M | 4M 37.3S | |||
| 3H 30M | 4M 58.6S | |||
| 3H 45M | 5M 19.9S | |||
| 4H 0M | 5M 41.2S | |||
| 4H 15M | 6M 2.6S | |||
| 4H 30M | 6M 23.9S | |||
| 4H 45M | 6M 45.2S | |||
| 5H 0M | 7M 6.6S | |||
| 5H 15M | 7M 27.9S | |||
| 5H 30M | 7M 49.2S | |||
| 5H 45M | 8M 10.6S | |||
| 6H 0M | 8M 31.9S |