Accelerate Bayesian analytics workflows in R through interactive modelling, visualization, and inference. Uses probabilistic graphical models as a unifying language for business stakeholders, statisticians, and programmers.
This package relies on the sleek and elegant greta package for
Bayesian inference. greta, in turn, is an interface into TensorFlow
from R. Future iterations of the causact package will aim to be a
front-end into several universal probablistic programming languages
(e.g. Stan, Turing, Gen, etc.).
Using the causact package for Bayesian inference is featured in The Business Analyst's Guide to Business Analytics available at
http://causact.com/.
NOTE: Package is under active development. Breaking changes are to be expected. Feedback and encouragement is appreciated via github issues or Twitter (https://twitter.com/preposterior).
You can install the current release version of the package from CRAN:
install.packages("causact")
or the development version from GitHub:
install.packages("remotes")
remotes::install_github("flyaflya/causact")
causact requires the greta package for Bayesian updating, which in
turn, requires a specific version of TensorFlow. Install both greta
and TensorFlow using the instructions available here:
http://causact.com/install-greta.html
Example taken from
https://www.causact.com/graphical-models-tell-joint-distribution-stories.html#graphical-models-tell-joint-distribution-stories
with the packages dag_foo() functions further described here:
library(causact)
graph = dag_create() %>%
dag_node(descr = "Get Card", label = "y",
rhs = bernoulli(theta),
data = carModelDF$getCard) %>%
dag_node(descr = "Card Probability", label = "theta",
rhs = beta(2,2),
child = "y") %>%
dag_plate(descr = "Car Model", label = "x",
data = carModelDF$carModel,
nodeLabels = "theta",
addDataNode = TRUE)
graph %>% dag_render()Hide model complexity, as appropriate, from domain experts and other less statistically minded stakeholders.
graph %>% dag_render(shortLabel = TRUE)library(greta)
#>
#> Attaching package: 'greta'
#> The following objects are masked from 'package:stats':
#>
#> binomial, cov2cor, poisson
#> The following objects are masked from 'package:base':
#>
#> %*%, apply, backsolve, beta, chol2inv, colMeans, colSums, diag,
#> eigen, forwardsolve, gamma, identity, rowMeans, rowSums, sweep,
#> tapply
gretaCode = graph %>% dag_greta(mcmc = FALSE)
#> ## The below greta code will return a posterior distribution
#> ## for the given DAG. Either copy and paste this code to use greta
#> ## directly, evaluate the output object using 'eval', or
#> ## or (preferably) use dag_greta(mcmc=TRUE) to return a data frame of
#> ## the posterior distribution:
#> y <- as_data(carModelDF$getCard) #DATA
#> x <- as.factor(carModelDF$carModel) #DIM
#> x_dim <- length(unique(x)) #DIM
#> theta <- beta(shape1 = 2, shape2 = 2, dim = x_dim) #PRIOR
#> distribution(y) <- bernoulli(prob = theta[x]) #LIKELIHOOD
#> gretaModel <- model(theta) #MODEL
#> draws <- mcmc(gretaModel) #POSTERIOR
#> drawsDF <- replaceLabels(draws) %>% as.matrix() %>%
#> dplyr::as_tibble() #POSTERIOR
#> tidyDrawsDF <- drawsDF %>% addPriorGroups() #POSTERIORlibrary(greta)
drawsDF = graph %>% dag_greta()
drawsDF ### see top of data frame
#> # A tibble: 4,000 x 4
#> theta_JpWrnglr theta_KiaForte theta_SbrOtbck theta_ToytCrll
#> <dbl> <dbl> <dbl> <dbl>
#> 1 0.831 0.251 0.610 0.188
#> 2 0.854 0.283 0.589 0.202
#> 3 0.868 0.252 0.626 0.199
#> 4 0.871 0.197 0.617 0.204
#> 5 0.876 0.260 0.654 0.188
#> 6 0.873 0.271 0.620 0.191
#> 7 0.859 0.242 0.610 0.210
#> 8 0.874 0.227 0.634 0.191
#> 9 0.874 0.227 0.634 0.191
#> 10 0.874 0.310 0.642 0.208
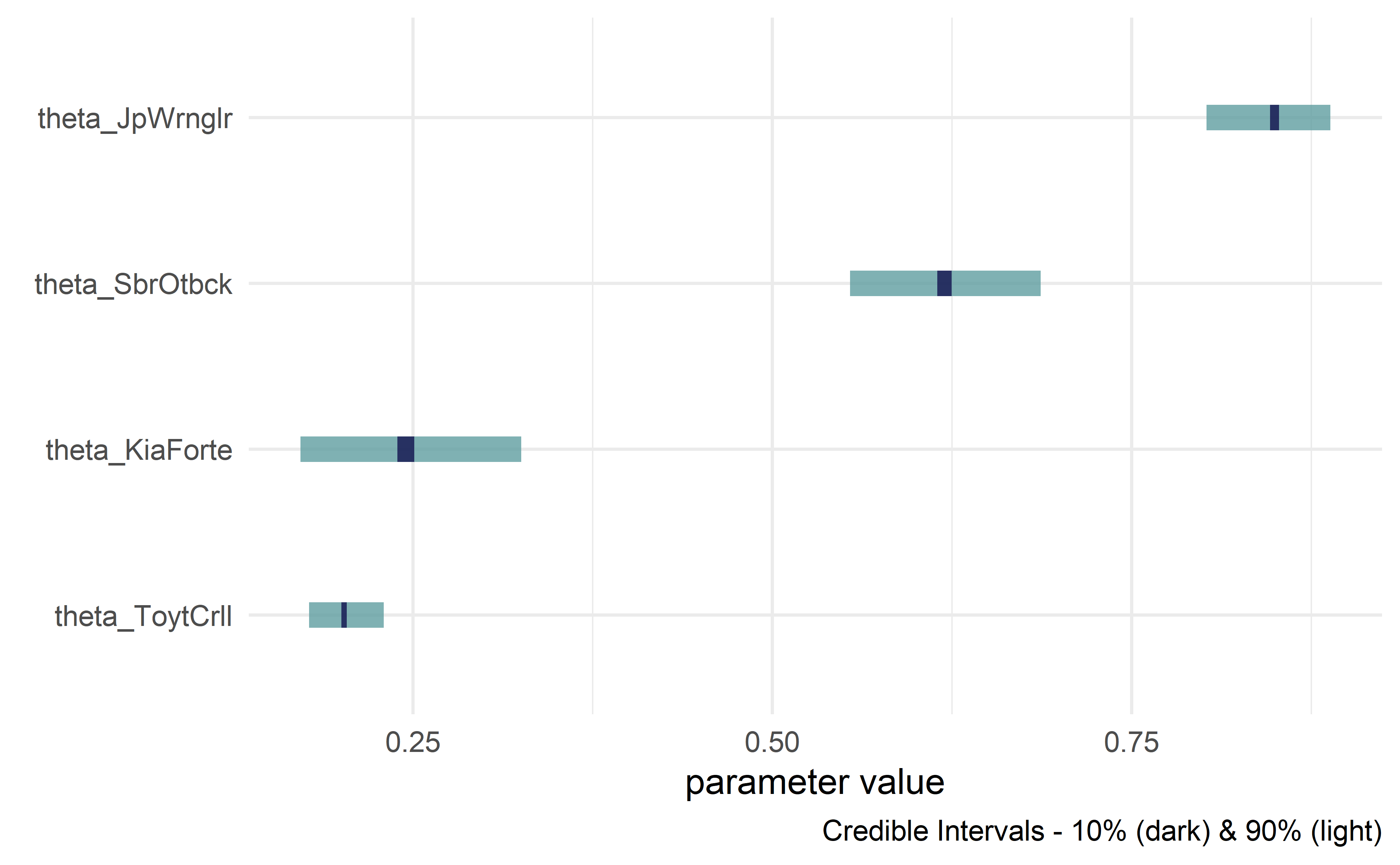
#> # ... with 3,990 more rowsdrawsDF %>% dagp_plot()For more info, see The Business Analyst's Guide to Business Analytics
available at https://www.causact.com. Two additional examples are
shown below.
McElreath, Richard. Statistical rethinking: A Bayesian course with examples in R and Stan. Chapman and Hall/CRC, 2018.
library(greta)
library(tidyverse)
library(causact)
# data object used below, chimpanzeesDF, is built-in to causact package
graph = dag_create() %>%
dag_node("Pull Left Handle","L",
rhs = bernoulli(p),
data = causact::chimpanzeesDF$pulled_left) %>%
dag_node("Probability of Pull", "p",
rhs = ilogit(alpha + gamma + beta),
child = "L") %>%
dag_node("Actor Intercept","alpha",
rhs = normal(alphaBar, sigma_alpha),
child = "p") %>%
dag_node("Block Intercept","gamma",
rhs = normal(0,sigma_gamma),
child = "p") %>%
dag_node("Treatment Intercept","beta",
rhs = normal(0,0.5),
child = "p") %>%
dag_node("Actor Population Intercept","alphaBar",
rhs = normal(0,1.5),
child = "alpha") %>%
dag_node("Actor Variation","sigma_alpha",
rhs = exponential(1),
child = "alpha") %>%
dag_node("Block Variation","sigma_gamma",
rhs = exponential(1),
child = "gamma") %>%
dag_plate("Observation","i",
nodeLabels = c("L","p")) %>%
dag_plate("Actor","act",
nodeLabels = c("alpha"),
data = chimpanzeesDF$actor,
addDataNode = TRUE) %>%
dag_plate("Block","blk",
nodeLabels = c("gamma"),
data = chimpanzeesDF$block,
addDataNode = TRUE) %>%
dag_plate("Treatment","trtmt",
nodeLabels = c("beta"),
data = chimpanzeesDF$treatment,
addDataNode = TRUE)graph %>% dag_render(width = 2000, height = 800)graph %>% dag_render(shortLabel = TRUE)drawsDF = graph %>% dag_greta()drawsDF %>% dagp_plot()Gelman, Andrew, Hal S. Stern, John B. Carlin, David B. Dunson, Aki Vehtari, and Donald B. Rubin. Bayesian data analysis. Chapman and Hall/CRC, 2013.
library(greta)
library(tidyverse)
library(causact)
# data object used below, schoolDF, is built-in to causact package
graph = dag_create() %>%
dag_node("Treatment Effect","y",
rhs = normal(theta, sigma),
data = causact::schoolsDF$y) %>%
dag_node("Std Error of Effect Estimates","sigma",
data = causact::schoolsDF$sigma,
child = "y") %>%
dag_node("Exp. Treatment Effect","theta",
child = "y",
rhs = avgEffect + schoolEffect) %>%
dag_node("Pop Treatment Effect","avgEffect",
child = "theta",
rhs = normal(0,30)) %>%
dag_node("School Level Effects","schoolEffect",
rhs = normal(0,30),
child = "theta") %>%
dag_plate("Observation","i",nodeLabels = c("sigma","y","theta")) %>%
dag_plate("School Name","school",
nodeLabels = "schoolEffect",
data = causact::schoolsDF$schoolName,
addDataNode = TRUE)graph %>% dag_render()drawsDF = graph %>% dag_greta()drawsDF %>% dagp_plot()