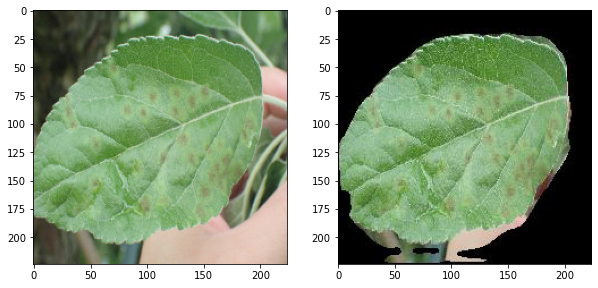
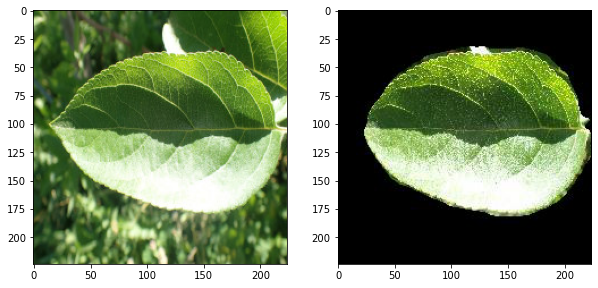
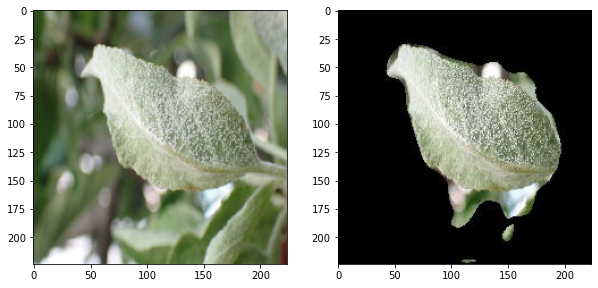
a unet model trained for the semantic segmentation of leaf images
.
├── dataset
│ └── note.md
├── model
│ ├── pretrained
│ │ └── download.md
│ ├── __init__.py
│ └── model.py
├── preprocess
│ ├── generate_dataset.py
│ ├── __init__.py
│ └── README.md
├── test
│ └── get_testset.sh
├── utilities
│ ├── __init__.py
│ └── utility.py
├── get_dataset.sh
├── get_pretrained.sh
├── LICENSE
├── predict.py
├── README.md
├── requirements.txt
└── train.py
input images are from the dataset of Plant Pathology 2021 Challenge
-
prepare dataset
linux users can run
get_dataset.shinstead of first three steps-
cd into
./preprocess -
download
DenseLeaves.zipfrom here -
unzip the downloaded file as
./preprocess/DenseLeaves/ -
run
python generate_dataset.py
The newly processed dataset is now saved at
./dataset -
-
cd back to project root and run
python train.pyto train the model
When training, the model saves the weights in the ./model/pretrained model.
latest_weights.pth- weights saved at the end of the last epochbest_val_weights.pth- weights saved when the model obtained minimum validation loss
- download pretrained weights
linux users can run get_pretrained.sh (make sure gdown is installed -- pip install gdown)
others can download the weights from links provided in this file
- specify test image location
edit the TEST_DIR variable in predict.py to specify custom images or download a sample dataset by running get_testset.sh in ./test folder.
tip: if the segmentation results are not satisfactory, modify the mask threshold values in
predict.pyfile
- DenseLeaves dataset - Michigan State University visit
- Plant Pathology 2021 dataset - Kaggle
2021-06-24: first code upload, most of the code is really bad (I wrote them a while ago). I shall refactor them soon.