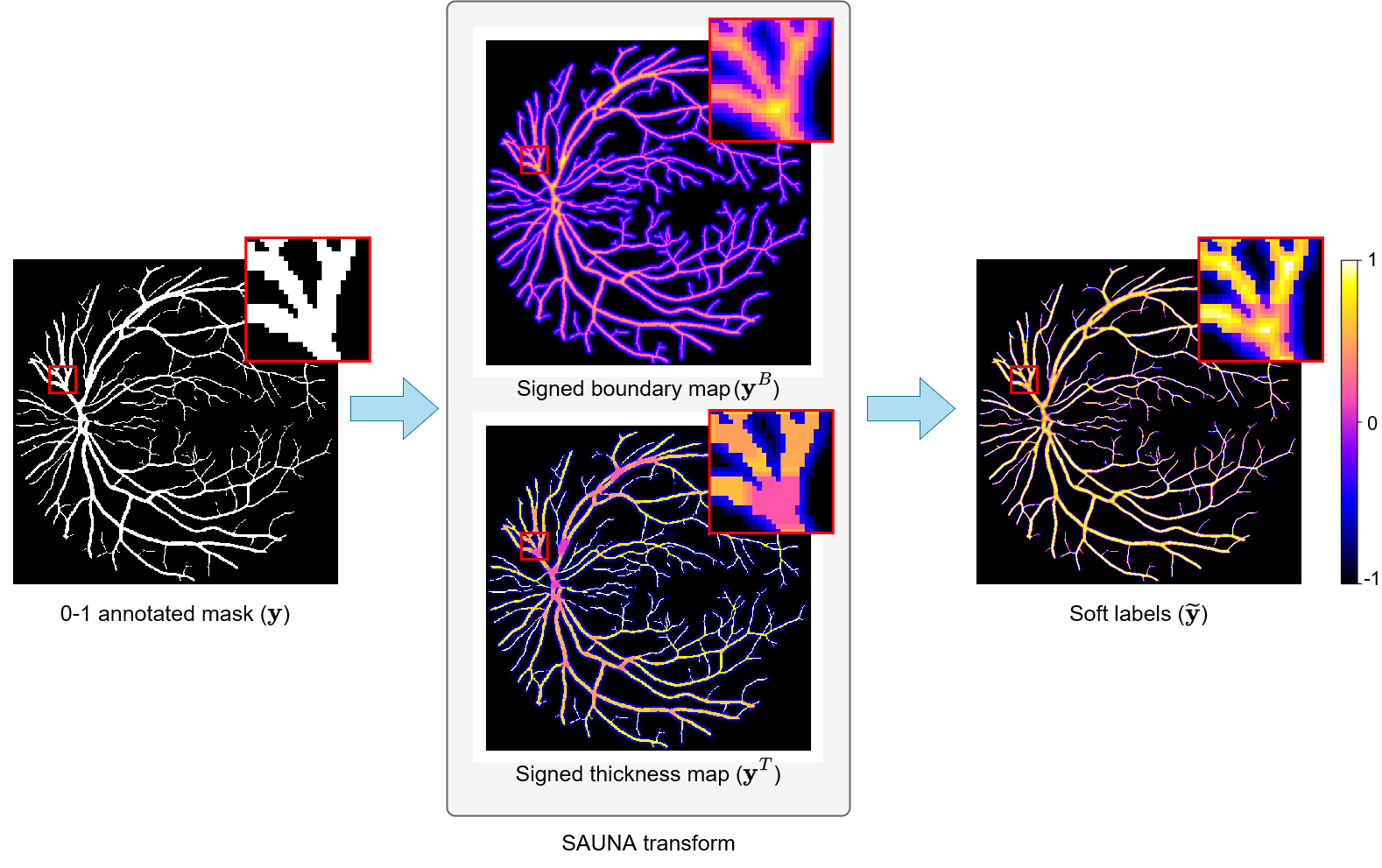
SAUNA: Image-level Regression for Uncertainty-aware Retinal Image Segmentation
We provide file env.yaml for dependencies.
conda env create -f env.yaml
conda activate sauna
pip install -e .cd mlpipeline/utils
python split_fives.pycd mlpipeline/utils
python ./generate_uncertainty_masks.py --root <ROOT_DIR> --in_dir <GT_DIR>The labels are available at URL.
python -m mlpipeline.train.run experiment=${EXP_NAME} \
model.params.cfg.arch=${ARCH_NAME}where
- EXP_NAME: experiment setting can be
fives_uncertainty_sem_seg(ours),fives_patch_sem_seg(for high-resolution-based methods), orfives_whole_sem_seg(for low-resolution-based methods). - ARCH_NAME: architecture name can be
Unet,UnetPlusPlus,IterNet,CTF-Net,CE-Net,DUnet,FR-Unet,DA-Net, orSwin-Unet.
python -m mlpipeline.train.evaluate \
--config=${EXP_NAME} \
--output_dir=/path/to/inference_results/${EXP_NAME} \
--log_dir=/path/to/eval/${EXP_NAME} \
--visual_dir=/path/to/visuals \
--metadata_path=/path/to/test_split.pkl \
--dataset_name=${DATASET} \
--seeds=${SEEDS} \
--folds=0,1,2,3,4where
- DATASET: is either
FIVES,STARE,DRIVE,CHASEDB1, orHRF.