Official PyTorch implementation of "Discovering Symbolic Expressions with Parallelized Tree Search"
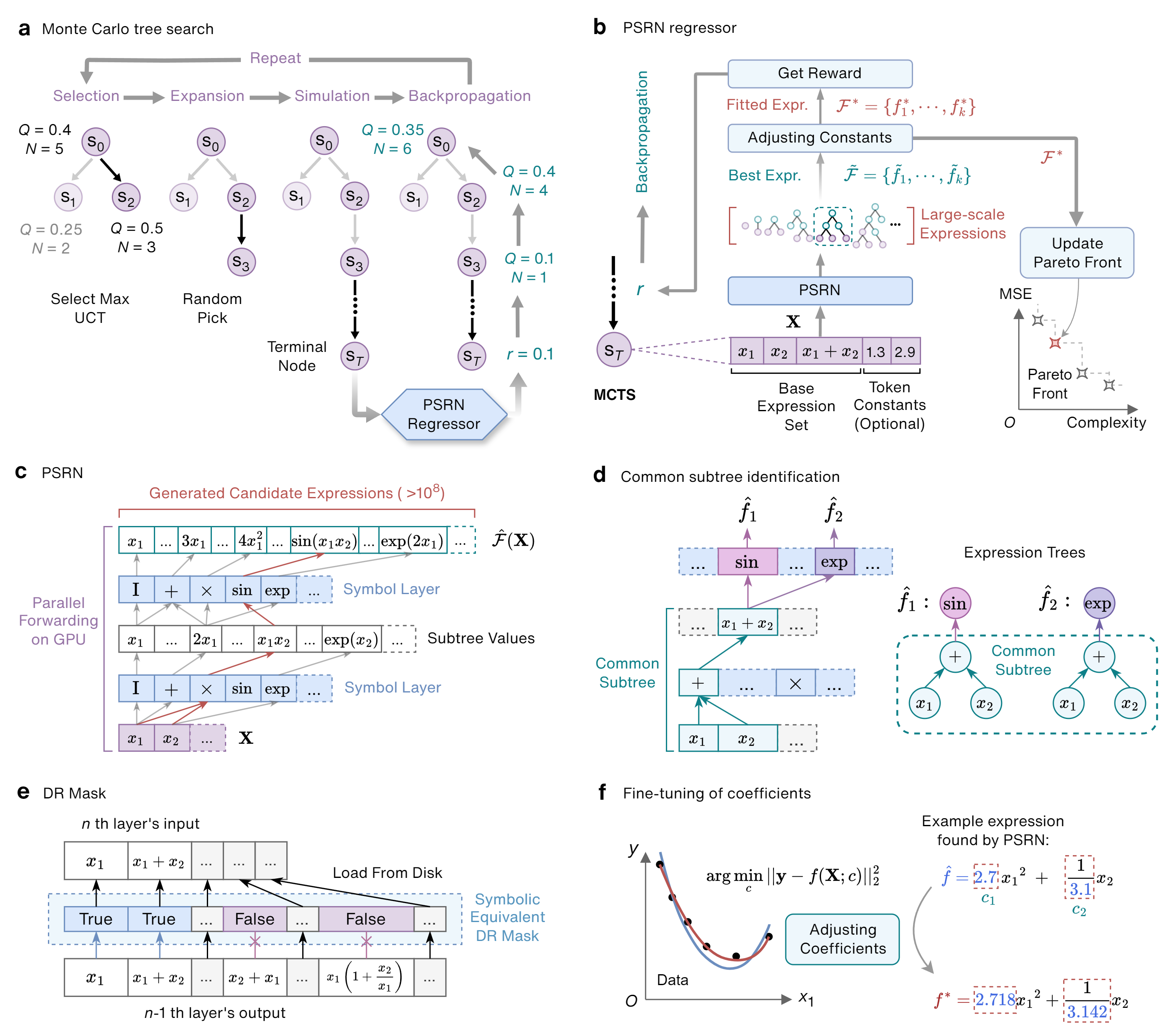
PTS (Parallelized Tree Search) with its core component PSRN (Parallel Symbolic Regression Network) is a novel symbolic regression framework featuring:
- Scalable: Evaluates hundreds of millions of candidate expressions within seconds
- Efficient: Automatically identifies and reuses common subtrees to avoid redundant computation
- Fast: Leverages GPU parallelization for massive expression evaluation
conda create -n PSRN python=3.8 "pytorch>=2.0.0" pytorch-cuda=12.1 -c pytorch -c nvidiaNote: Adjust the pytorch-cuda version as necessary based on your GPU's CUDA version.
conda activate PSRN
pip install pandas==1.5.3 click==8.0.4 dysts==0.1 numpy==1.22.3 scipy==1.7.3 tqdm==4.65.0 pysindy==1.7.5 derivative==0.6.0 scikit-learn==1.3.0 sympy==1.10.1Notes:
- If using a version of PyTorch below 2.0, an error may occur during the
torch.topkoperation. - The experiments were performed on servers with Nvidia A100 (80GB) and Intel(R) Xeon(R) Platinum 8380 cpus @ 2.30GHz.
- We recommend using a high-memory GPU as smaller cards may encounter CUDA memory errors under our experimental settings. If you experience memory issues, consider reducing the number of input slots or opting for
semi_kozaoperator sets (e.g., replacing"Sub"and"Div"with"SemiSub"and"SemiDiv") orbasicoperator sets (e.g., replacing"Sub"and"Div"with"Neg"and"Inv").
To execute the script with custom data, use the following arguments:
-g: Specifies the GPU to use. Enter the GPU index.-i: Sets the number of input slots for PSRN.-c: Indicates whether to include constants in the computation (True / False).-l: Defines the operator library to be used. Specify the name of the library or an operator list.--csvpath: Specifies the path to the CSV file to be used. By default, if not specified, it uses./custom_data.csv. Each column represents an independent variable.
For more detailed parameter settings, please refer to the run_custom_data.py script.
To run the script with custom data with an expression probe (the algorithm will stop when it finds the expression or its symbolic equivalents), use:
python run_custom_data.py -g 0 -i 5 -c False --probe "(exp(x)-exp(-x))/2"Without an expression probe, use:
python run_custom_data.py -g 0 -i 5 -c FalseTo activate 2 constant tokens during each forward pass in PSRN, enter:
python run_custom_data.py -g 0 -i 5 -c True -n 2 --probe "(exp(x)-exp(-x))/2"In case of limited VRAM (or the ground truth expression is expected to be simple), consider reducing the input size with this command:
python run_custom_data.py -g 0 -i 2 -c False --probe "(exp(x)-exp(-x))/2"To customize the operator library, you can specify it like so (may need to generate dr_mask first):
python run_custom_data.py -g 0 -i 5 -c False --probe "(exp(x)-exp(-x))/2" -l "['Add','Mul','Identity','Tanh','Abs']"For custom data paths, specify the CSV path as follows:
python run_custom_data.py -g 0 -i 5 -c False --probe "(exp(x)-exp(-x))/2" --csvpath ./another_custom_data.csvThe .npy files under ./dr_mask are pre-generated. When you try to use a new network architecture (e.g., a new combination of operators, number of variables, and number of layers), you may need to run the gen_dr_mask.py script first. Typically, this process takes less than a minute.
For example:
python utils/gen_dr_mask.py --n_symbol_layers=3 --n_inputs=5 --ops="['Add','Mul','SemiSub','SemiDiv','Identity','Sin','Cos','Exp','Log','Tanh','Cosh','Abs','Sign']"To reproduce our experiments, execute the following command:
python run_benchmark_all.py --n_runs 100 -g 0 -l koza -i 5 -b benchmark.csvFor the Feynman expressions:
python run_benchmark_all.py --n_runs 100 -g 0 -l semi_koza -i 6 -b benchmark_Feynman.csvThe Pareto optimal expressions and corresponding statistics for each puzzle are available in the log/benchmark directory. Additionally, the expected runtime for each puzzle can be found in the supplementary materials.
Discovering the dynamics of chaotic systems by running the following command
python run_chaotic.py --n_runs 50 -g 0 # Using GPU index 0This script will generate Pareto optimal expressions for each derivative, and the outcomes will be stored in the log/chaotic directory.
Then, you can assess the symbolic recovery rate by executing:
python result_analyze_chaotic.pyThis analysis will automatically compute and save the statistics to log/chaotic_symbolic_recovery/psrn_stats.csv
python run_realworld_EMPS.py --n_runs 20 -g 0 # Using GPU index 0The results (Pareto optimal expressions) can be found in log/EMPS
python run_realworld_roughpipe.py --n_runs 20 -g 0 # Using GPU index 0The results (Pareto optimal expressions) can be found in log/roughpipe
To reproduce our ablation studies, execute the following command.
The results will be stored in the log/ directory.
python study_ablation/mcts/run_random.py -x ablation_mcts --n_runs 100 -g 0 -l koza -i 5 -r False
python study_ablation/mcts/run_random.py -x ablation_mcts --n_runs 100 -g 0 -l koza -i 5 -r Truepython study_ablation/constants/run_c_experiments.py --n_runs 20 -g 0 You can modify the operator library using the -l flag, adjust the number of input slots with -i, and choose whether to use the DR Mask.
While the script is running, monitor the memory footprint using nvidia-smi or nvitop.
python study_ablation/drmask/run_without_drmask.py --use_drmask True -i 4 -l koza -g 0
python study_ablation/drmask/run_without_drmask.py --use_drmask False -i 4 -l koza -g 0python study_ablation/noise/run_noise.py --experiment_name=noise --n_runs 100 -g 0 -l arithmetic -b benchmark_noise.csvIf you use this work, please cite:
@article{arxiv:2407.04405,
author = {Ruan, Kai and Gao, Ze-Feng and Guo, Yike and Sun, Hao and Wen, Ji-Rong and Liu, Yang},
title = {Discovering symbolic expressions with parallelized tree search},
journal = {arXiv preprint arXiv:2407.04405},
year = {2024},
url = {https://arxiv.org/abs/2407.04405}
}