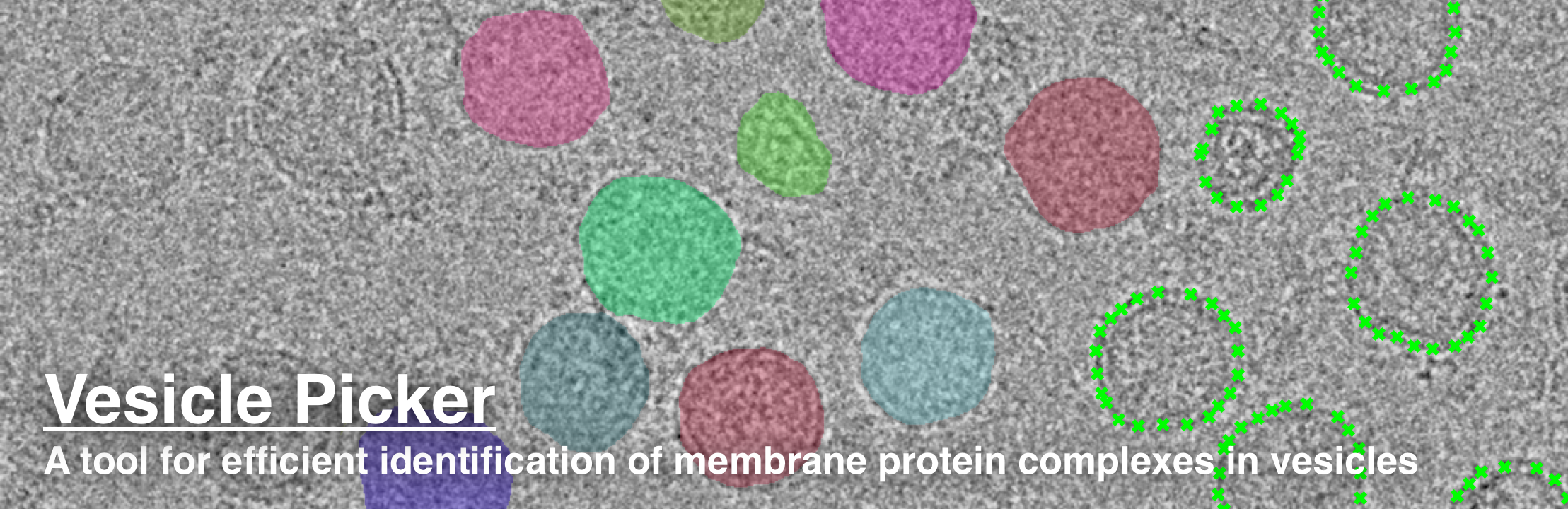
-
Ensure Git and Anaconda (or Miniconda) are installed on your computer.
-
If you wish to run the Segment Anything model on GPU, ensure CUDA is installed on your machine. CUDA is not necessary to run Segment Anything on your computer's CPU, but the program will run much slower.
-
Clone the Vesicle Picker repository:
git clone https://github.com/r-karimi/vesicle-picker.git -
Enter this repository:
cd vesicle-picker -
Create a clean conda virtual environment.
conda create -n vesicle-picker conda activate vesicle-picker conda install pip -
Edit the
pyproject.tomlfile in the base directory to install the correction version of PyTorch, PyTorch vision, and PyTorch audio for your computer. These instructions differ based on whether you are installing PyTorch for CPU or GPU usage.-
Visit the PyTorch installation page and select the appropriate options, ensuring that Pip is selected as the package manager and CPU is selected as the compute platform. Note the given install command, but do not run it.
-
Modify
install-pytorchinpyproject.tomlwith the install command noted above:# Example: CPU install-pytorch = "pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cpu"
-
Note your version of CUDA and Python by running:
nvcc --version python --version -
Browse the PyTorch wheels to find the appropriate versions of PyTorch, PyTorch vision, and PyTorch audio for your installed versions of CUDA and Python (e.g.
cu118for CUDA 11.8 andcp39for Python 3.9). -
Modify the
install-pytorchcommand inpyproject.tomlto match these versions:# Example: Python 3.9.X and CUDA 11.8 install-pytorch = "pip install torch==2.1.1+cu118 torchvision==0.16.1+cu118 torchaudio==2.1.1+cu118 -f https://download.pytorch.org/whl/torch_stable.html"
-
-
Install
vesicle-pickerand dependencies:pip install . poe install-pytorch -
Download the Segment Anything model weights from the Segment Anything GitHub page and place them in the
vesicle-pickerrepository. We recommend trying with the ViT-L model weights first. -
Modify csparc_login.ini to match the active cryoSPARC instance from which micrographs will be imported into Vesicle Picker and into which particle locations will be exported.
Vesicle Picker is broadly subdivided into three sub-programs, each of which can be run from a seperate script. These sub-programs are find_vesicles.py, filter_vesicles.py, and generate_picks.py. The scripts can be run from the command line and take one argument, which is the path of the respective parameter file for a script. This parameter file is where one can set all the necessary parameters to execute a sub-program. For instance, the parameters for filter_vesicles sub-program consist of general parameters such as pixel size and cryoSPARC instance information, as well as specific parameters such as the minimum area or roundness for a predicted vesicle area to be selected for subsequent analysis.
Before processing your own dataset, we recommend working through the introductory Jupyter notebook find_vesicles.ipynb. This notebook describes how the program imports data residing in cryoSPARC and describes each step of the processing pipeline. It also allows for empirical fine-tuning of the various parameters mentioned above on a test image from a dataset. We note in our paper that a combination of roundness and area postprocessing filters are sufficient to obtain high precision and recall in the task of finding synaptic vesicles. If these are sufficient for your dataset as well, then the parameters that need to be set by the user are as follows.
-
$\sigma_{space}$ ,$\sigma_{colour}$ , and$d$ for the bilateral filter. Increasing these parameters blurs the image such that Vesicle Picker can operate at higher recall at the expense of precision. -
$Roundness_{min}$ for the roundness filter. The roundness ranges from 0 (i.e. the object is a line) to 1 (i.e. the object is a perfect circle) for every predicted vesicle. -
$Area_{min}$ and$Area_{max}$ for the area filter. This parameter is useful if a user knows the rough size of the vesicles in advance. -
$r_{dilation}$ or$r_{erosion}$ for particle picks offset from the membrane edge. This parameter can be useful if the user has prior knowledge about the position of proteins of interest relative to the lipid bilayer. - Box size to control the density of the picks. A higher density of picks can lead to an increased likelihood of identifing a protein of interest because one of the picks is more likely to be well-centered on the protein. However, this can come at the cost of increased computational burden as the number of particle images subjected to 2D classification is increased.
There are a variety of other postprocessing filters that can be applied to your data as well. These filters are commented out in parameters/filter_vesicles.ini by default, but can be uncommented and applied depending on the specific use case of the user. More information about the various postprocessing methods implemented in this library can be found in vesicle_picker/postprocessing.py.
To process your own full dataset with the command line scripts, follow the steps below:
-
Import your movies, then perform patch motion correction and patch CTF estimation. Vesicle Picker operates on motion corrected micrographs.
-
Curate your motion corrected micrographs to discard any micrographs that, for instance, contain no vesicles. Vesicle Picker is designed to operate on the output of a Curate Exposures job.
-
Note the project ID, workspace ID, and job ID of your Curate Exposures job. You will input this information into the parameter files to run the sub-programs referenced below.
-
Find vesicles by modifying the
find_vesicles.iniparameter file with your desired parameters, ensuring you fill in the correct cryoSPARC information. Also make sure to fill in your cryoSPARC login information, usingcsparc_login.inias a template. Finally, indicate an appropriate output directory for the detected vesicles. The detected vesicles will be stored as Python.pklfiles in the output directory. We have pre-filled this parameter file with a reasonable set of starting parameters.When you're ready, run the
find_vesicles.pyscript. The script takes a file path to the parameters file as its only argument:python find_vesicles.py parameters/find_vesicles.ini -
Filter the found vesicles by modifying the
filter_vesicles.iniparameter file, uncommenting the selection filters that you want to use and setting their minimum and maximum values. Ensure you set the input directory for this script as the output directory offind_vesicles.py. Again, we have pre-filled this parameter file with a reasonable set of starting parameters.When you're ready, run
filter_vesicles.py:python filter_vesicles.py parameters/filter_vesicles.ini -
Generate particle picks by modifying the
generate_picks.iniparameter file. Set the workspace into which the vesicle picks will be exported. Set the dilation or erosion radius if desired, and set box size parameter to control the density of picks. We recommend picking with a reasonably high density (i.e. a box size less than half of the expected diameter of your protein of interest) and removing duplicate particles later in cryoSPARC. Ensure you set the input directory for this script as the output directory offilter_vesicles.py.Run
generate_picks.py:python generate_picks.py parameters/generate_picks.iniOnce this script has finished, you should see a collection of
.pklfiles in the output directory of this script, as well as a new, completed job in cryoSPARC called Vesicle Picks. This job will be used for downstream processing in cryoSPARC.
-
Extract particles from the micrographs that were used as input to
find_vesicles.py. We recommend extracting with a box size 2x to 3x larger than the box size used to generate picks. -
Proceed with downstream analysis in cryoSPARC, such as 2D classification and Ab initio reconstruction.
-
We recommend experimenting with different model architectures and downsampling factors to find a good trade-off between accuracy and speed when processing a full dataset. We have found that perfect recall when finding vesicles is usually unnecessary for obtaining a structure. A small set of high-quality vesicles are usually more informative than vesicles mixed with non-vesicle objects, so do not be afraid of stringently filtering your vesicles.
-
If you are able to generate good 2D classes of a membrane protein complex with Vesicle Picker, these particles can be used for template matching and training a Topaz model to obtain a larger and better centered particle stack for subsequent 3D reconstruction and refinement.
-
When performing 2D classification, particularly when searching for small membrane proteins and protein complexes, we found that it is important to perform at least 40 iterations of expectation-maximization. We also typically increase the batchsize per class to 150 or 200. Finally, we almost always see better results when we disable the
Recenter 2D classesparameter. -
We typically iterate 2D classification and selection of promising 2D classes several times. In early iterations, we enable the
Force Max over poses/shiftsparameter to efficiently classify large numbers of particles. In later iterations, where images of proteins in membranes are enriched, we typical disable theForce Max over poses/shiftsparameter to better resolve low SNR particles within membranes. -
Sometimes, a user will already have a high quality particle stack and is interested in filtering an existing stack by selecting particles on vesicles found by vesicle picker. If this is the case, we recommend exporting a set of dense (i.e. picked with a small box size) vesicle picks to cryoSPARC and using the
Remove Duplicatesjob, to intersect the two existing particle stacks. See this job's cryoSPARC documentation for more details.
| Failure Mode | Suggestion |
|---|---|
| Model is not detecting vesicles. | Increase the filter diameter and |
| Model detects vesicles and annotates patches in the background of the image. | Decrease the filter diameter and |
| Model cannot discriminate between vesicles and contaminants. | Experiment with the available filters. If vesicles are larger than contaminants on average, increase the minimum area filter. If contaminants are irregularly shaped, increase the minimum roundness filter. |
Karimi, R., Coupland, C. E. & Rubinstein, J. L. Vesicle Picker: A tool for efficient identification of membrane protein complexes in vesicles. bioRxiv 2024.07.15.603622 (2024) doi:10.1101/2024.07.15.603622.