Profile HMM demo code and data
- clustal-omega-1.2.3
- argtable2-13
- hmmer-3.1b2-macosx-intel
- Java and python environment
- Jnetpcap
- Install HMMER 3
- Install clustal-omega-1.2.3
- Try data from folder Data using scripts from folder called Shellscript
- Put the test data and the script in the same folder
- run ./buildmodel.sh (it will cost some time)
- Read result
This folder contains programs to crawl and collect pcapfiles.
This folder contains some key research data for reproducing.
This folder includes pcap file processing code.
This folder contains modified KFP algorithm, and sample testing data. And srcipts for analyses.
This folder contains shellsripts to analyze gene sequence file data and scripts to parse results.
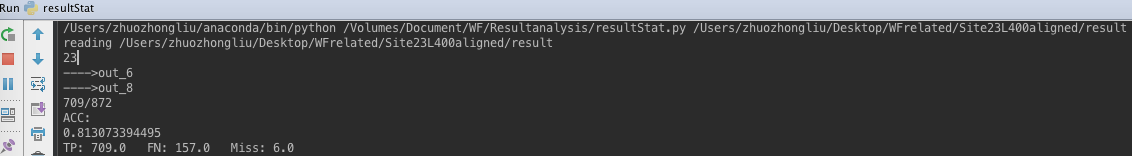
- PHMM result for Alexa23 L400 after MSA alignment.
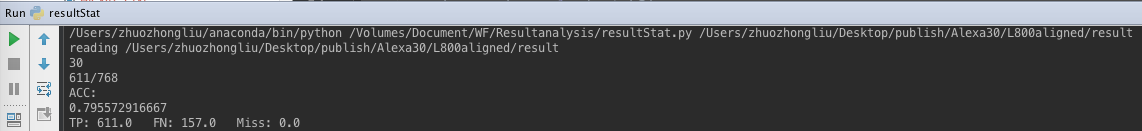
- PHMM result for Alexa30 L800 after MSA alignment.
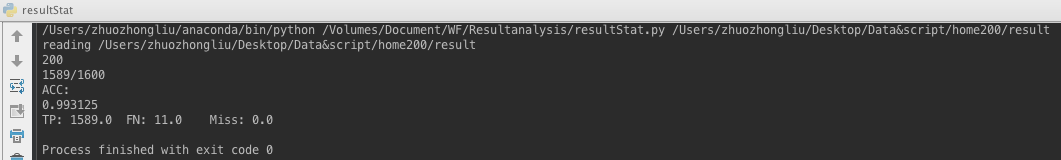
- PHMM result for Alexa200 L400 after MSA alignment.
- Full sequence test result for Alexa30 using model L=800..