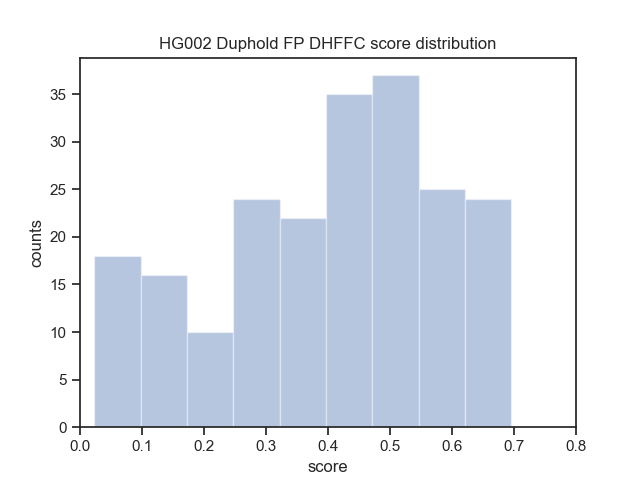
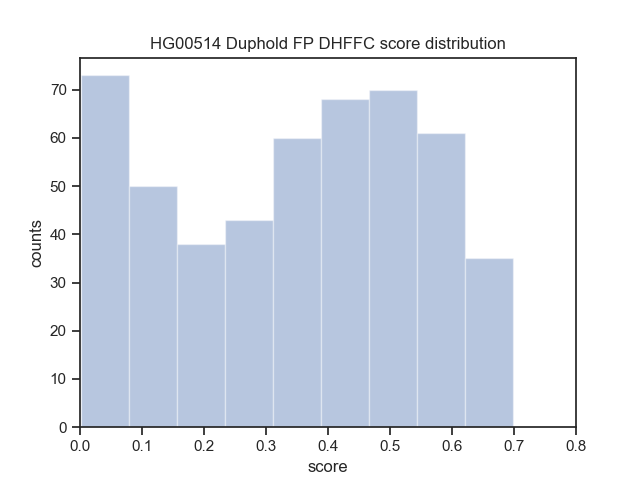
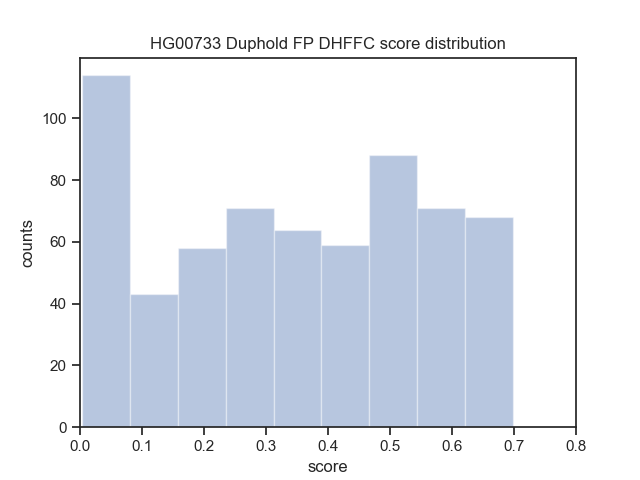
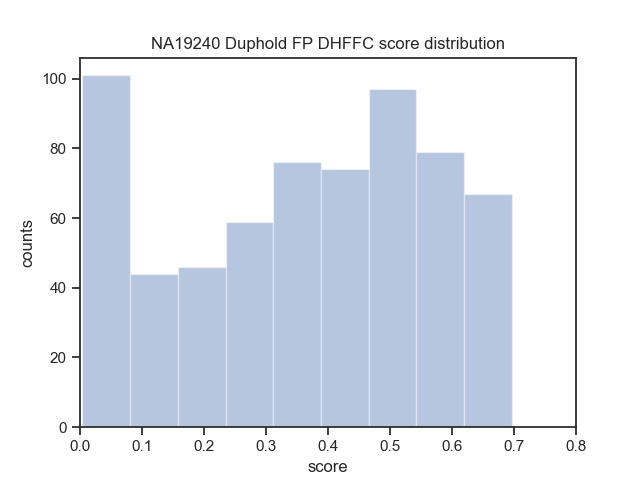
Download the Cram indices (.crai) listed on the high_coverage index ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000_genomes_project/1000genomes.high_coverage.GRCh38DH.alignment.index
- Aligned to GRCh38 reference genome
bcftools view -i 'SVTYPE="DEL"' $VCF \
| bcftools query -f '%CHROM\t%POS\t%INFO/END[\t%SAMPLE,%GT]\n' \
| python sample_del.py > $OUT/del.sample.bed # from git repo- Output to bed file annotated with sample and genotype (REF, HET, ALT)
- TODO make the variables command line args with flags
cat $BED_FILE | gargs -p $PROCESSES \
"bash gen_img.sh \\
--chrom {0} --start {1} --end {2} --sample {3} --genotype {4} \\
--fasta $FASTA \\
--bam-list $CRAM_LIST \\
--bam-dir $CRAM_INDEX_DIR \\
--out-dir $OUT_DIR/imgs"- Use gargs to parse the contents of the Training regions and feed to
gen_img.sh $CRAM_LIST(they’re actually crams) can be absolute file paths or urls (must download indices though)$CRAM_DIRis the directory containing the CRAM indices- TODO upload the cram list of urls to github
- TODO check if I used a different GRCh38 from “full_analysis_set_plus_decoy_hla”
bash crop.sh \
--processes $NUM_PROCESSES \
--data-dir $DATA_DIR- Where
$DATA_DIRis the parent directory containing the img/ directory from the previous step - Cropped images will be placed in
$DATA_DIR/crop
- TODO
We use the run.py script to train a new model
usage: run.py train [-h] [--batch-size BATCH_SIZE] [--epochs EPOCHS]
[--model-type MODEL_TYPE] --data-dir DATA_DIR
[--learning-rate LR] [--momentum MOMENTUM]
[--label-smoothing LABEL_SMOOTHING] [--save-to SAVE_TO]
optional arguments:
-h, --help show this help message and exit
--batch-size BATCH_SIZE, -b BATCH_SIZE
Number of images to feed to model at a time. (default:
80)
--epochs EPOCHS, -e EPOCHS
Max number of epochs to train model. (default: 100)
--model-type MODEL_TYPE, -mt MODEL_TYPE
Type of model to train. (default: CNN)
--data-dir DATA_DIR, -d DATA_DIR
Root directory of the training data. (default: None)
--learning-rate LR, -lr LR
Learning rate for optimizer. (default: 0.0001)
--momentum MOMENTUM, -mom MOMENTUM
Momentum term in SGD optimizer. (default: 0.9)
--label-smoothing LABEL_SMOOTHING, -ls LABEL_SMOOTHING
Strength of label smoothing (0-1). (default: 0.0)
--save-to SAVE_TO, -s SAVE_TO
filename if you want to save your trained model.
(default: None)
- TODO
- TODO
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/NIST_SVs_Integration_v0.6/HG002_SVs_Tier1_v0.6.vcf.gz ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/NIST_SVs_Integration_v0.6/HG002_SVs_Tier1_v0.6.bed
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/hgsv_sv_discovery/data/CHS/HG00514/high_cov_alignment/
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/hgsv_sv_discovery/data/PUR/HG00733/high_cov_alignment/
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/hgsv_sv_discovery/data/YRI/NA19240/high_cov_alignment/
ftp://ftp.ncbi.nlm.nih.gov/pub/dbVar/data/Homo_sapiens/by_study/genotype/nstd152
- Remove length 0 contigs (causes problems with truvari otherwise)
- Run
fix_vcf.pyscript to correct SVLEN- For some reason the %INFO/END field is just start + 1 so we need to use SVLEN to calculate the true end.
bcftools view -i 'SVTYPE="DEL"' $TRUTH_SET_VCF \
| grep -v "length=0>" \
| python fix_vcf.py \
| bgzip -c > $FIXED_TRUTH_SET
tabix $FIXED_TRUTH_SETUse the following command
smoove call \
--outdir $OUT_DIR \
--processes $PROCESSES \
--name $SAMPLE_NAME \ # eg HG00514
--exclude $BED_DIR/exclude.cnvnator_100bp.GRCh38.20170403.bed
--fasta $FASTA #
--removepr \
--support $SUPPORT \
--genotype \
--duphold \
$CRAM_PATHYou can get the exclude regions bed for GRCh38 from here
fasta index
s3://1000genomes/technical/reference/GRCh38_reference_genome/GRCh38_full_analysis_set_plus_decoy_hla.fa s3://1000genomes/technical/reference/GRCh38_reference_genome/GRCh38_full_analysis_set_plus_decoy_hla.fai
bcftools view -i 'SVTYPE="DEL"' $SAMPLE-smoove.genotyped.vcf.gz \
| bgzip -c > $SAMPLE-smoove.genotyped.del.vcf.gzbcftools query -f '%CHROM\t%POS\t%INFO/END[\t%SAMPLE\t%GT]\n' \
$SAMPLE-smoove.genotyped.del.vcf.gz | gargs -p $PROCESSES \
"bash gen_img.sh \\
--chrom {0} --start {1} --end {2} --sample $SAMPLE --genotype DEL \\
--fasta $FASTA \\
--bam-dir $PATH_TO_CRAM \\
--out-dir $OUT_DIR/imgs"bash crop.sh \
--processes $NUM_PROCESSES \
--data-dir $DATA_DIR- Where
$DATA_DIRis the parent directory containing the img/ directory from the previous step - Cropped images will be placed in
$DATA_DIR/crop
cd $SAMPLE_DIR # parent directory of cropped images
find $(pwd)/crop/*.png > $IMAGE_LISTbcftools view -i 'DHFFC<0.7' $BASELINE_VCF | bgzip -c > dhffc.lt.0.7.vcf.gz
tabix dhffc.lt.0.7.vcf.gzbash create_test_vcfs.sh \
--model-path $MODEL_PATH \
--data-list $IMAGE_LIST \
--vcf $BASELINE_VCF \ # i.e. the smoove genotyped vcf
--out-dir $OUT_DIRbash truvari.sh \
--comp-vcf $COMP_VCF \
--base-vcf $TRUTH_SET_VCF \
--reference $REF \
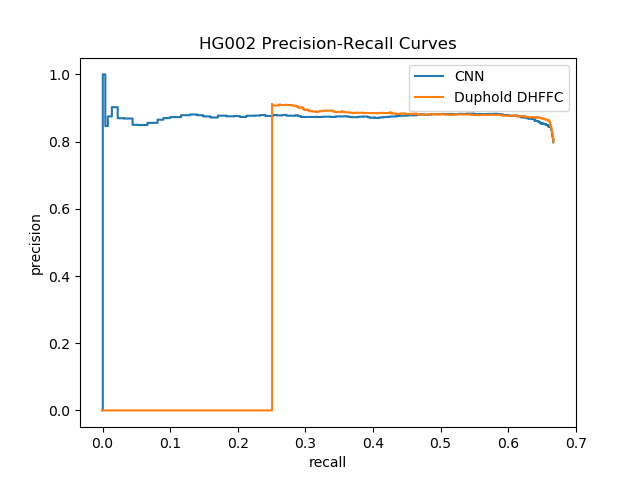
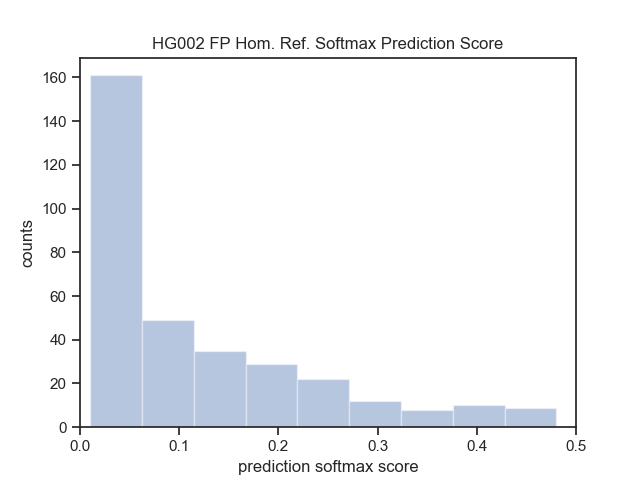
--out-dir $OUT_DIR| HG002 (Ashkenazim) with tier 1 | Smoove | DHFFC | CNN | manta | DHFFC | CNN |
|---|---|---|---|---|---|---|
| TP | 1496 | 1488 | 1489 | |||
| FP | 83 | 33 | 62 | |||
| FN | 276 | 284 | 283 | |||
| Precision | 0.947 | 0.978 | 0.96 | |||
| Recall | 0.844 | 0.840 | 0.840 | |||
| F1 | 0.893 | 0.904 | 0.896 | |||
| FP Intersection | ||||||
| HG002 (Ashkenazim) without tier 1 | Smoove | DHFFC | CNN | manta | DHFFC | CNN |
| TP | 1787 | 1764 | 1758 | 1708 | 1687 | 1706 |
| FP | 452 | 276 | 273 | 265 | 175 | 187 |
| FN | 893 | 916 | 922 | 972 | 993 | 981 |
| Precision | 0.798 | 0.865 | 0.866 | 0.866 | 0.906 | 0.901 |
| Recall | 0.667 | 0.658 | 0.656 | 0.637 | 0.629 | 0.634 |
| F1 | 0.727 | 0.747 | 0.746 | 0.734 | 0.7428 | 0.744 |
| FP Intersection |
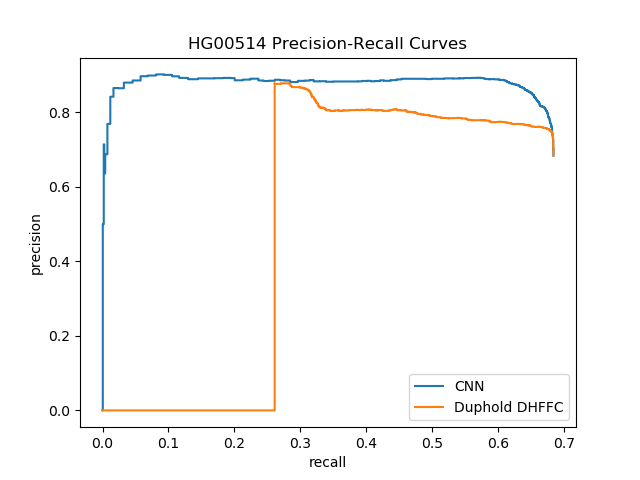
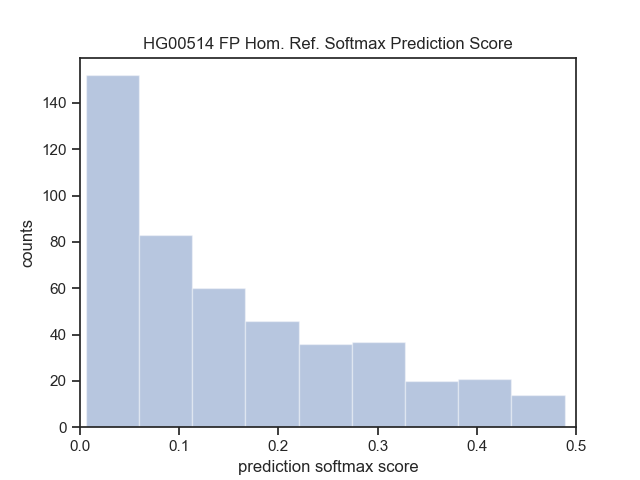
| HG00514 (Han Chinese) | Lumpy | DHFFC | CNN | manta | DHFFC | CNN |
|---|---|---|---|---|---|---|
| TP | 1860 | 1837 | 1803 | 1779 | 1759 | 1751 |
| FP | 860 | 596 | 372 | 502 | 328 | 221 |
| FN | 858 | 881 | 915 | 939 | 959 | 967 |
| Precision | 0.684 | 0.755 | 0.829 | 0.780 | 0.843 | 0.888 |
| Recall | 0.684 | 0.676 | 0.663 | 0.654 | 0.647 | 0.644 |
| F1 | 0.684 | 0.713 | 0.737 | 0.712 | 0.731 | 0.747 |
| HG00514 (Sensitive) | Lumpy | DHFFC | CNN | |||
| TP | 1875 | 1851 | 1814 | |||
| FP | 1157 | 761 | 477 | |||
| FN | 843 | 867 | 904 | |||
| Precision | 0.618 | 0.709 | 0.792 | |||
| Recall | 0.690 | 0.681 | 0.667 | |||
| F1 | 0.652 | 0.695 | 0.724 |
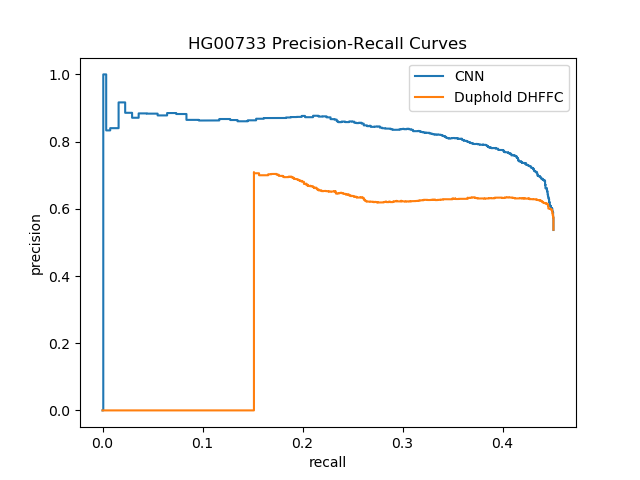
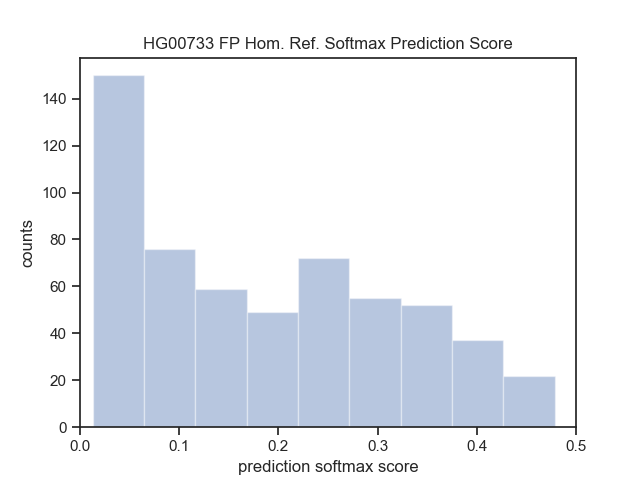
| HG00733 (Puerto Rican) | Smoove | DHFFC | CNN | manta | DHFFC | CNN |
|---|---|---|---|---|---|---|
| TP | 1236 | 1216 | 1181 | 1774 | 1753 | 1736 |
| FP | 1066 | 760 | 517 | 455 | 306 | 204 |
| FN | 1505 | 1525 | 1560 | 967 | 988 | 1005 |
| Precision | 0.537 | 0.615 | 0.696 | 0.796 | 0.851 | 0.895 |
| Recall | 0.451 | 0.443 | 0.431 | 0.647 | 0.640 | 0.633 |
| F1 | 0.490 | 0.520 | 0.532 | 0.714 | 0.730 | 0.742 |
| HG00733 (Sensitive) | Lumpy | DHFFC | CNN | |||
| TP | 1277 | 1255 | 1219 | |||
| FP | 1422 | 968 | 676 | |||
| FN | 1464 | 1486 | 1522 | |||
| Precision | 0.473 | 0.564 | 0.643 | |||
| Recall | 0.466 | 0.460 | 0.445 | |||
| F1 | 0.469 | 0.506 | 0.526 | |||
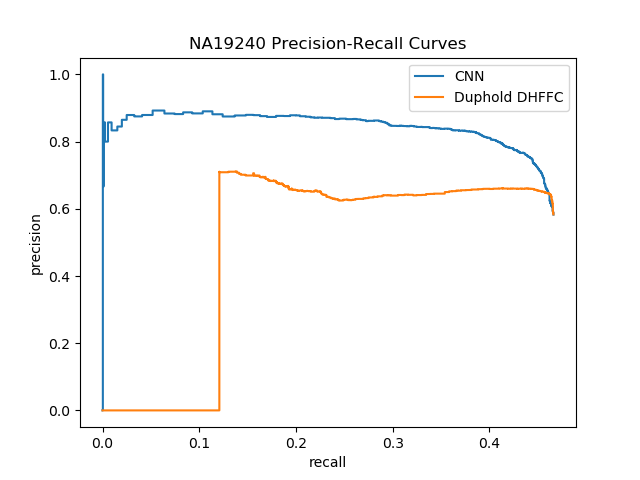
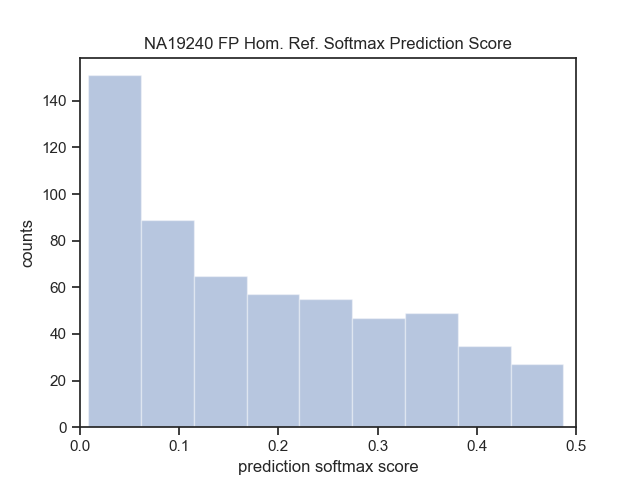
| NA19240 (Yoruban) | Lumpy | DHFFC | CNN | manta | DHFFC | CNN |
| TP | 1494 | 1470 | 1414 | 2067 | 2054 | 2019 |
| FP | 1070 | 801 | 628 | 520 | 359 | 272 |
| FN | 1711 | 1735 | 1791 | 1138 | 1151 | 1186 |
| Precision | 0.583 | 0.647 | 0.692 | 0.799 | 0.851 | 0.881 |
| Recall | 0.566 | 0.459 | 0.441 | 0.645 | 0.641 | 0.630 |
| F1 | 0.518 | 0.537 | 0.549 | 0.714 | 0.731 | 0.735 |
| NA19240 (Sensitive) | Lumpy | DHFFC | CNN | |||
| TP | 1542 | 1518 | 1466 | |||
| FP | 1427 | 1023 | 804 | |||
| FN | 1663 | 1687 | 1739 | |||
| Precision | 0.519 | 0.597 | 0.646 | |||
| Recall | 0.481 | 0.474 | 0.457 | |||
| F1 | 0.500 | 0.528 | 0.536 |
- TODO write this up in more detail
- ie. data sources, and how to generate images, etc.
bedtools intersect -wb -f 1.0 -r -a $svplaudit_bed -b $pred_bed \
| python3 svplaudit_agreement.py- For regions with SV plaudit scores < 0.2 and > 0.8 we have ~97% agreement
- If we don’t filter out unambiguous regions then we have ~93% agreement.
bcftools view -i 'SVTYPE="DEL"' $NA12878_callset \
| bcftools query -f '%CHROM\t%POS\t%INFO/END\n' \
| bedtools intersect -wb -f 1.0 -r -a stdin -b $pred_bed \
| python3 vcf_agreement.py- For the original callset we have ~93% agreement.
- Can this be rectified?
- Just analyze the fn’s made by duphold/CNN but not smoove
- ie. does duphold/CNN make largely the same or different mistakes
- Just make a representative sample of true positives
- Again, for fn’s just do the ones not made by smoove
- if yes then do it. (it can’t)