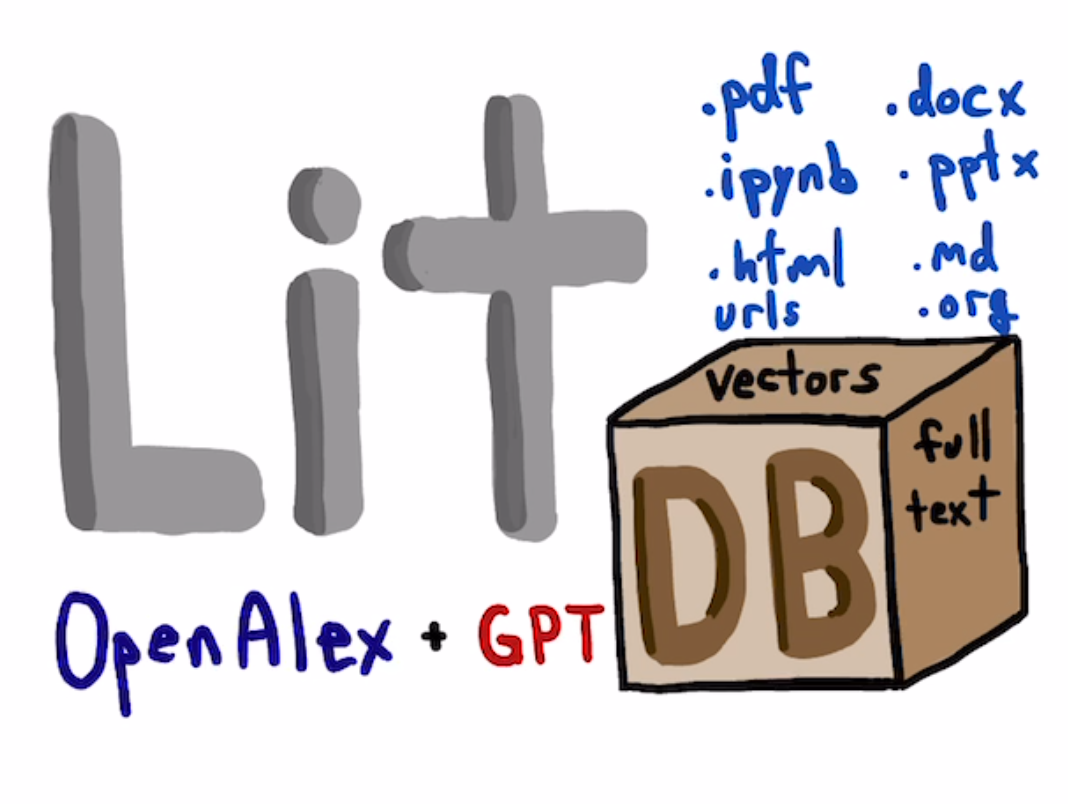
litdb is a tool to help you curate and use your collection of scientific literature. You use it to collect and search papers. You can use it to collect older articles, and to keep up with newer articles. litdb uses https://openalex.org for searching the scientific literature, and https://turso.tech/libsql to store results in a local database.
The idea is you add papers to your database, and then you can search it with natural language queries, and interact with it via an ollama GPT application. It will show you the papers that best match your query. You can read those papers, get bibtex entries for them, or add new papers based on the references, papers that cite that paper, or related papers. You can also set up filters that you update when you want to get new papers created since the last time you checked.
- https://www.youtube.com/live/e-J3Bh2Uti4 Introduction to litdb
- https://www.youtube.com/live/teW68WogulU local files (volume is very low for some reason)
- https://youtube.com/live/3LltpiiQaR8 CrossRef, reviewer suggestions, COA
- https://youtube.com/live/ZkKKuvVUWkE litdb and Emacs
- https://youtube.com/live/j7rItPwWDaY litdb and Jupyter Lab
- https://youtube.com/live/SUtvtc7l6y0 litdb + GPT enhancements
- https://youtube.com/live/3FZ1ROnCC6Y litdb + LiteLLM and streamlit
- https://www.youtube.com/live/IKKTQSTXQmc litdb + Youtube and audio
- https://youtube.com/live/MEf9rPI0Z1M litdb + Image search with text and image queries using CLIP
- https://youtube.com/live/C4qCam0shf8 litdb + deep research
- https://youtube.com/live/6Wpy7KM3wIM litdb + LLM-augmented search of OpenAlex
- https://www.youtube.com/live/eS0D-Aje_6A litdb + Claude Desktop
litdb is on PyPi.
pip install litdbTo get the cutting edge package, you can install it directly from GitHUB.
pip install git+https://github.com/jkitchin/litdblitdb relies on a lot of ML-related packages, and conflicts with version are common. If you have any issues, I recommend you install it with uv (https://github.com/astral-sh/uv) like this:
uv venv --python 3.13
source .venv/bin/activate
uv pip install litdbYou have to activate that virtual env to use litdb, which may be annoying.
You have to create a toml configuration file. This file is called litdb.toml. The directory this file is in is considered the root directory. All commands will start in the current working directory and look up to find this file. You can put this file in your home directory, or you can have sub-directories, e.g. a per project litdb.
There are a few choices you have to make. You have to choose a SentenceTransformer model, and specify the size of the vectors it makes. You also have to specify the chunk_size and chunk_overlap settings that are used to break documents up to compute document level embedding vectors.
You will need an OpenAlex premium key if you want to use the update-filters feature.
[embedding] model = 'all-MiniLM-L6-v2' cross-encoder = 'cross-encoder/ms-marco-MiniLM-L-6-v2' chunk_size = 1000 chunk_overlap = 200 [openalex] email = "[email protected]" api_key = "..." [gpt] model = "llama2" [llm] model = "ollama/llama2"
You can define an environment variable to the root of your default litdb project. This should be a directory with a litdb.toml file in it.
export LITDB_ROOT="/path/to/your/default/litdb"When you run a litdb command, it will look for a dominating litdb.toml file, which means you are running the command in a litdb project. If one is not found, it will check for the LITDB_ROOT environment variable and use that if it is found. Finally, if that does not exist, it will prompt you to make a new project in the current directory.
Your litdb starts out empty. You have to add articles that are relevant to you. It is an open question of the best way to build a litdb. The answer surely depends on what your aim is. You have to compromise on breadth and depth with the database size. The CLI makes it pretty easy to do this
litdb has a cli with an entry command of litdb and subcommands (like git) for interacting with it. You can see all the options with this command.
litdb --helpYou have to start somewhere. You can use this to open a search in OpenAlex.
litdb web queryYou can also open searches with these options:
| option | source |
|---|---|
| -g, –google | |
| -gs, –google-scholar | Google Scholar |
| -ar, –arxiv | Arxiv |
| -pm, –pubmed | Pubmed |
| -cr, –chemrxiv | ChemRxiv |
| -br, –biorxiv | BioRxiv |
| -a, –all | All |
You can find starting points this way.
This is a default query in Open Alex. It does not change your litdb, it just does a simple text search query on works.
litdb openalex queryYou can get more specific with a filter:
litdb openalex -f 'author.orcid:https://orcid.org/0000-0003-2625-9232'You can also search other endpoints and use fulters. Here we perform a search on Sources for display_names that contain the word discovery.
litdb openalex -e sources -f display_name.search:discoveryThe previous command lets you define a precise keyword or filter search for OpenAlex. An alternative is the lsearch sub-command. This command uses a LLM to generate a set of keyword searches for OpenAlex. Then, for each keyword search it returns results that are sorted in ascending and descending citation counts, publication year, and it performs a random sample search. Each result is scored by vector similarity to the original query, and the top results are printed.
litdb lsearch "some query of interest"some options include:
- -q
- integer count of number of keyword queries to generate
- -n
- integer count of results to retrieve for each keyword query
- -k
- integer count of how many results to display
You add an article by its DOI. There are optional arguments to also add references, citing and related articles.
litdb add doi --references --citing --relatedTo add an author, use their orcid. You can use litdb author-search firstname lastname to find an orcid for a person.
litdb add orcidTo add entries from a bibtex file, use the path to the file.
litdb add /path/to/bibtex.bibYou can provide more than one source and even mix them like this.
litdb add doi1 doi2 orcidThese are all one-time additions.
You can also add things like YouTube videos and podcasts. We use ML to extract the audio from these to text so they become searchable!
litdb provides several convenient ways to add queries to update your litdb in the future.
To get new papers by an author, you can follow them.
litdb follow orcidlitdb watch "filter to query"litdb citing doilitdb related doiA filter is used in OpenAlex to search for relevant articles. Here is an example of adding a filter for articles in the journal Digital Discovery. This doesn’t add any entries directly, it simply stores the filter in the database. The main difference of this vs the watch command above is the explicit description.
litdb add-filter "primary_location.source.id:https://openalex.org/S4210202120" -d "Digital Discovery"You can get a list of your filters like this.
litdb list-filtersYou can update the filters like this.
litdb update-filtersThis adds papers that have been created since the last time you ran the filter. You need an OpenAlex premium API key for this. This will update the last_updated field.
You can remove a filter like this:
litdb rm-filter "filter-string"I find it helpful to review your litdb. To get a list of articles added in the last week, you can run this command.
litdb review -s "1 week ago"This works best when you update your litdb regularly. You might want to redirect that into a file so you can review it in an editor of your choice.
There are several search options.
The main way litdb was designed to be searched is with by natural language queries. The way this works is your query is converted to a vector using SentenceTransformers, and then a vector search identifies entries in the database that are similar to your query.
litdb vsearch "natural language query"The default number of entries returned is 3. You can change that with an optional argument
litdb vsearch "natural language query" -n 5There is an iterative version of vsearch called isearch. This finds the closest entries, then downloads the citations, references and related entries for each one, and repeats the query until you tell it to stop, or it doesn’t find any new results.
litdb isearch "some query"There is a full text search (full on the text in litdb) available. The command looks like this.
litdb fulltext "query"See https://sqlite.org/fts5.html for information on what the query might look like. The search is done with this SQL command:
select source, text from fulltext where text match ? order by rankThe default number of entries returned is 3. You can change that with an optional argument
litdb fulltext "natural language query" -n 5Vector and full text search have complementary strengths and weaknesses. We combine them in the hybrid-search subcommand. This performs two searches on two different queries, and combines them with a unified score that is used to rank all the matches. This ensures you get some results that match the full search, and the vector search. It is worth trying if you aren’t finding what you want by vector or text search alone.
litdb hybrid-search "vector query" "text query"You can use litdb as a RAG source for ollama. This looks up the three most related papers to your query, and uses them as context in a prompt to ollama (with the llama2 model). I find this quite slow (it can be minutes to generate a response on an old Intel Mac). I also find it makes up things like references, and that it is usually necessary to actually read the three papers. The three papers come from the same vector search described above.
litdb gpt "what is the state of the art in automated laboratories for soft materials"litdb supports litellm so you can use almost any LLM provider you want: OpenAI, Anthropic, Gemini, whatever you have an API key for.
The free tier of the API includes 1,500 requests per day with Gemini 1.5 Flash.
It uses a different command than the ollama gpt command.
litdb chat "what is the state of the art in automated laboratories for soft materials"There are some fancy things you can do with the prompt:
- Avoid using RAG if –norag is in your prompt.
- If you surround python objects with backticks, it will try expanding that to the documentation from Python.
- A line that starts with < indicates a shell command to run and the output will be expanded into the prompt.
- A prompt of !save will save the current chat to a file.
- You can use this syntax to expand a file or url in the prompt for context:
[[file/url]]
Your prompt history is saved in your litdb, so you can go back to them if you want.
If you prefer a browser, you can now launch a streamlit app for litdb:
litdb appThis should launch the app in your browser and you can search litdb from it. The terminal application is more advanced in terms of prompt expansion.
This command will record audio, transcribe that audio to text, and then do a vector search on that text. You will be prompted when the recording starts, and you press return to stop it. litdb will show you what it heard, and ask if you want to do a vector search on it.
litdb audio -pI haven’t found the transcription to be that good on technical scientific terms. This is a proof of concept capability.
Note that you need to install these libraries for this feature to work:
pyaudio, playsound, SpeechRecognition
These are not trivial to install, and pyaudio relies on external libraries like portaudio that may not be easy to install. These are currently commented out in pyproject.toml because of these difficulties.
You can copy a screenshot to the clipboard, and then use OCR to extract text from it, and do a vector search on that text.
litdb screenshotIf you can copy and paste text, you should do that instead. This is helpful to get text from images, or pdfs where the text is stored in an image, maybe from videos, or screen share from online meetings, etc.
Eventually, if images get integrated into litdb, this is also an entry point for image searches.
litdb supports tagging entries so you can group them. To tag a source with tag1 and tag2, use this syntax.
litdb add-tag source -t tag1 -t tag2You can remove tags like this.
litdb rm-tag source -t tag1 -t tag2You can delete a tag from the database.
litdb delete-tag tag1To see all the tags do this.
litdb list-tagsTo see entries with a tag:
litdb show-tag tag1You can use this to export tagged entries into bibtex entries like this.
litdb show-tag workflow -f '{{ source }}' | litdb bibtexYou can use these commands to export bibtex entries or citation strings.
This command will try to generate a bibtex entry for entries in your litdb.
litdb bibtex doi1 doi2The output can be redirected to a file.
You can also use a search like this and pipe the output to litdb bibtex.
litdb vsearch "machine learning in catalysis
" -f "{{ source }}" | litdb bibtexThis command will output a citation for the sources. It is mostly a convenience function. There is not currently a way to customize the citation.
litdb citation doi1 doi2You can also use a search like this and pipe the output to litdb bibtex.
litdb vsearch "machine learning in catalysis
" -f "{{ source }}" | litdb citationsYou can use litdb to find freely available PDFs via https://unpaywall.org/.
litdb unpaywall doiThese do not always work, and sometimes you get a version from arxiv or pubmed.
litdb is just a sqlite database (although you need to use the libsql executable for vector search). There is a CLI way to run a sql command. For example, to find all entries with a null bibtex field and their types use a query like this.
litdb sql "select source, json_extract(extra, '$.type'), json_extract(extra, '$.bibtex') as bt from sources where bt is null"You might also use this for very specific queries. For example, here I search the citation strings for my name.
litdb sql "select source, json_extract(extra, '$.citation') as citation from sources where citation like '%kitchin%'"Deep research is one of the newer ideas in LLMs. It is a combination of web and other searches to find relevant information, coupled with an LLM to synthesize that information into a textual response. Litdb supports deep research with your database using gpt-researcher with this command:
litdb research "some query"litdb will generate some documents from your database from your query, and use this in a research task with gpt_researcher. This takes longer than a simple chat, but is often much richer in detail, and summarizes across documents.
The default setup is to use a local ollama model, with arxiv as a search tool. You will get better results most likely if you use a cloud LLM. To use these you can add this new section to litdb.toml:
[gpt-researcher]
FAST_LLM = "google_genai:gemini-2.0-flash"
SMART_LLM = "google_genai:gemini-2.0-flash"
STRATEGIC_LLM = "google_genai:gemini-2.0-flash"
EMBEDDING = "google_genai:models/text-embedding-004"
See https://docs.gptr.dev/docs/gpt-researcher/llms/llms for other configurations.
The default search engine in litdb for deep research is DuckDuckGo and arxiv. If you setup these API keys you can also use pubmed (https://www.ncbi.nlm.nih.gov/account/), google (https://developers.google.com/custom-search/v1/overview), and Tavily for search.
export NCBI_API_KEY=
export GOOGLE_CX_KEY=
export TAVILY_API_KEY=I have not tried the other retrievers at https://github.com/assafelovic/gpt-researcher/tree/master/gpt_researcher/retrievers.
You can do research on local documents with this command:
litdb research "some query" --doc-path=/path/to/documentsIt is not currently possible to use litdb do research with and use a local directory full of documents at the same time.
The default is to print the report to stdout. To write to a file instead use:
litdb research "some query" -o report.mdYou specify other output formats, anything pandoc can convert to. However, I found pdf a challenge for pypandoc, and use md2pdf for that instead. On Mac, if you see errors related to gobject, you probably have do run something like this to fix it.
brew list pango gobject-introspection
export DYLD_LIBRARY_PATH=$(brew --prefix)/lib:$DYLD_LIBRARY_PATHlitdb_mcp install /path/to/litdb.libsqlThen, restart Claude Desktop.
Now in the Claude Desktop app you can use prompts like “Search my litdb for references on alloy segregation” or “Search OpenAlex for articles on alloy segregation.”
The idea of using local files is that it is likely you have collected information in the form of files on your hard drive, and you want to be able to find information in those files.
It is possible to add any file that can be turned into text to litdb. That includes:
- docx
- pptx
- html
- ipynb
- org / md
- bib
- url
This limits portability because you need a path if you want to be able to open that file.
The same vector, fulltext and gpt search commands are available for local file entries. These tend to be longer documents than the OpenAlex entries, and I am not sure how well the search works at the document level embeddings. Search at a chunk level is very precise; odds are you want paragraph level similarity to your query.
An early version of litdb stored each chunk. This is possible, but I used another table for it. You could munge the source to be something like f.pdf::chunk-1 so each one is unique, but that seems more complicated and you would need to do some experiments to see if it is warranted.
You can combine this with the OpenAlex entries in a single database.
You can walk a directory and add files from it with this command.
litdb index dir1This directory is saved and you can update all the previously indexed directories like this.
litdb reindexSome annoying things that may happen are duplicate content, e.g. because you have the same file in multiple formats like docx and pdf, or because you have literal copies of files in multiple places.
You should also be careful sharing a litdb that has indexed local files. It may have sensitive information that you don’t want others to be able to find.
Of course there is some Emacs integration. I made a new link for litdb.
litdb:https://doi.org/10.1021/jp047349j
The links export as \cite{source}, and there is a function litdb-generate-bibtex to export bibtex entries for all links in the buffer. These entries are not certain to be valid, most likely from the keys (some DOIs are probably invalid keys).
You can easily insert a link like this:
M-x litdb
See ./litdb.el for details. This is not a package on MELPA yet. You should just load the .el file in your config. You can also use litdb-fulltext, litdb-vsearch, and litdb-gpt from Emacs to interact with your litdb.
litdb.el is under active development, and will be an alternative UI to the terminal eventually. It is too early to tell if it will replace org-ref. It has potential, but that would be a very large undertaking.
litdb uses a sqlite database with libsql. libsql is a sqlite fork with additional capabilities, most notably integrated vector search.
The main table in litdb is called sources.
- sources
- source (url to source location)
- text (the text for the source)
- extra (json data)
- embedding (float32 blob in bytes)
- date_added string
This table has an embedding_idx index for vector search.
There is also a virtual table fulltext for fulltext search.
- fulltext
- source
- text
And a table called queries.
- queries
- filter
- description
- last_updated
This database is automatically created when you use litdb.
The text that is stored for each entry comes from OpenAlex and is typically limited to the title and abstract. For the text in each entry The first line is typically a citation including the title, and the rest is the abstract if there is one. I feel like I see more and more entries with no abstract. This will certainly limit the quality of search, and could bias results towards entries with more text in them.
The quality of the vector search depends on several things. First, litdb stores a document level embedding vector that is computed by averaging the embedding vectors of overlapping chunks. We use Sentence Transformers to compute these. There are many choices to make on the model, and these have not been tested exhaustively. So far ‘all-MiniLM-L6-v2’ works well enough. There are other models you could consider like getting embeddings from ollama, but at the moment litdb can only use SentenceTransformers.
I guess that document level embeddings are less effective on longer documents. The title+abstract from OpenAlex is pretty short, and so far there isn’t evidence this is a problem.
Second, we rely on defaults in libsql for the vector search, notably finding the top k nearest vectors based on cosine similarity. There are other distance metrics you could use like L2, but we have not considered these.
The query is based on vector similarity between your query and the texts. So, you should write the query so it looks like what you want to find, rather than as a question. It is less clear how you should structure your query if you are using the GPT capability. It is more natural to ask a question, or give instructions. The RAG is still done by similarity though.
Finally, the search can only find things that are in your database. If you haven’t added it there, you won’t find it. That definitely means you will miss some papers. I try to use a mesh of approaches to cover the most likely papers. This includes:
- Follow authors
- add references, related, and citing papers to the most relevant papers.
- Use text search filters
- Add papers I find from X, bluesky, LinkedIn, etc. (and their references, related, etc)
- If read a paper in litdb that is good, add its references, related, etc.
It is an iterative process, and you have to make a judgment call about when to stop it. You can always come back later. There might even be newer papers to find.
Similar limitations exist for local files. There are additionally the following known limitations:
- The quality of document to text influences the ultimate embedding. This varies by type of document, and the library used to convert it.
- Local files tend to be longer documents and this can lead to hundreds of text chunks per document. These chunk embeddings are averaged into one embedding. It is not obvious this is as effective as vector search on each chunk, but it is more memory efficient.
For PDF to text we use pymupd4fll which works for this proof of concept. There is a Pro version of that package which supports more file formats. It is not obvious what it would cost to use that. I used docling in an early prototype. It also worked pretty well, but it was a little slower I think, and would occasionally segfault so I stopped using it. Spacy is integrating PDF to structured data using docling (https://explosion.ai/blog/pdfs-nlp-structured-data). There is plenty of room for improvement in this dimension, with trade offs in performance and accuracy.
There is a new package from Microsoft to convert Office files to Markdown (https://github.com/microsoft/markitdown) that they specifically mention using in the context of LLMs.
The embedding model we use is trained on text. It is probably not as good at finding code, and the gpt we use is also probably not good at generating code. I guess you would need another table in the database for code, and a different model for embedding and generation. This only matters if you index jupyter notebooks (and later if other code files are supported).
Vector search is the core requirement for litdb. There are many ways to achieve this. I only considered local solutions so the options are:
- sqlite + vectorlite (https://github.com/1yefuwang1/vectorlite)
- sqlite + sqlite-vec (https://github.com/asg017/sqlite-vec)
- libsql https://github.com/tursodatabase/libsql
vectorlite aims to be faster than sqlite-vec, but it relies on hnsw for vector search, and I was uncomfortable figuring out how to set the size of the db for this application.
sqlite-vec is nice, and early versions of litdb used it and its precursor. This approach requires a virtual table for the embeddings. This is installed as an extension, and is still considered in early stages of adoption.
libsql is a fork of sqlite with integrated vector search, and potential for using it as a cloud database. It is supported by a company, with freemium cloud services. In libsql you store the vectors in a regular table, and search on an embedding index. The code is on GitHUB, and can also be used locally.
These are ideas for future expansion.
I am not sure what the best way to do this is. The records in litdb are stored by the source, often a url, or path. The PDFs would be stored outside the database, and we would need some way to link them. The keys aren’t suitable for naming, but maybe a hash of the keys would be suitable. This would add a fuller opportunity to search larger, local documents too. In org-ref, I only had one pdf per entry. I guess here I would have a new table, so you could have multiple documents linked to an entry, although it won’t be easy to tell what they are from the hash-based filenames.
Notes on the other hand, might be small enough that they could be stored in the database. Then they would be easily searchable. They could also be stored externally to make them easy to edit. I haven’t found the notes feature in org-ref that helpful, and usually I take notes in various places. What I should do is add a search to find the litdb links in your org-files. This is already a feature of org-db.
An alternative to the CLI and Emacs would be to run this in Jupyter Lab with magic commands and rich output.
It might be helpful to have a graph representation of a paper that shows nodes of citing, references, and related papers, with edge length related to a similarity score, and node size related to number of citations.
ResearchRabbit and Litmaps do this pretty well.
There might be a way to get better results using agents and / or tools. For example, you might have a tool that can lookup new articles on OpenAlex, or augment with google search somehow. Or there might be some iterative prompt building tool that refines the search for related articles based on output results.
Here are some references for when I get back to this.
- https://github.com/ollama/ollama-python
- https://github.com/MikeyBeez/Ollama_Agents
- https://github.com/premthomas/Ollama-and-Agents
- https://medium.com/@lifanov.a.v/integrating-langgraph-with-ollama-for-advanced-llm-applications-d6c10262dafa
- https://medium.com/@abhilasha.sinha/building-a-crew-of-agents-with-open-source-llm-using-ollama-to-analyze-fund-documents-as-multi-page-756d8fd9fbf0
- https://blog.paperspace.com/building-local-ai-agents-a-guide-to-langgraph-ai-agents-and-ollama/
I don’t use llamaindex (maybe I should see what it does), but it has this section on agents https://docs.llamaindex.ai/en/stable/understanding/agent/
It might be nice to have a flask app with an API. This would facilitate interaction with Emacs.
Almost everything is done synchronously and it blocks the program. At least some things could be done asynchronously I think, and that might speed things up (especially for local files), or at least let you do other things while it happens.
The only thing to be careful about is not exceeding rate limits to OpenAlex. This is handled in the synchronous code.
I use a generic embedding model, and there are others that are better suited for specific tasks. For example:
- MatBERT cite:&trewartha-2022-quant-advan
- Scibert cite:&beltagy-2019-sciber
- Matscibert cite:&gupta-2022-matsc
- Specter cite:&cohan-2020-spect https://www.sbert.net/docs/sentence_transformer/pretrained_models.html#scientific-similarity-models
- PaECTER cite:&ghosh-2024-paect for patents
These might have a variety of uses with litdb that range from extracting data, named entity recognition, specific searches on materials, etc.
It is not essential to use SentenceTransformers for embedding, they are just easy to use. An alternative is something like ollama embeddings (https://ollama.com/blog/embedding-models) or llama.cpp https://github.com/abetlen/llama-cpp-python?tab=readme-ov-file#embeddings. The main reason to use on of these would be performance, and maybe better integration with a chat llm.
It is not that easy to just switch models; you would need to either add new columns and compute embeddings for everything, or update all the embeddings for a new model. The SPECTER embedding is much bigger than the all-MiniLM-L6-v2 embedding.
from sentence_transformers import SentenceTransformer
m = SentenceTransformer('allenai-specter')
print(m.encode(['test']).shape)
I have setup litdb to be project based. There may come a time when it is desirable to merge some set of databases. It might not be necessary, I think you can attach databases in sqlite (https://www.sqlitetutorial.net/sqlite-attach-database/) to achieve basically the same effect. litdb doesn’t store version info at the moment, so it could be tricky to ensure compatibility.
Still it might be interesting to sync two databases, e.g. https://www.sqlite.org/rsync.html. I don’t know if this works with libsql, but it might allow there to be a central db that users pull from.
The first version of litdb with libsql used a fully remote db on their cloud. The main benefit of that is you can update the db from another machine, keeping your working machine load low. A secondary benefit would be using the db from different machines more easily. Right now I use Dropbox to sync it; that mostly works but I get some conflict files here and there if I change it on one machine while it is open on another machine. It is a little more complex to set up though, and I got several api errors on long running scripts, and with network issues, so I switched to this local setup. I think you could specify this in the litdb.toml file and have it do the right thing on a project basis.
One day it might be possible to include images in this (https://www.sbert.net/docs/sentence_transformer/pretrained_models.html#image-text-models). At the moment, OpenAlex entries do not have any images, but other web resources and local files could. I have an image database in org-db, but I don’t use it a lot.
Vector search might miss some things. Full text search is hard to do with meaning. There are several ways to do a hybrid search, e.g. do a full text search on keywords, and a vector search, and use some kind of union on those results.
https://www.meilisearch.com/blog/full-text-search-vs-vector-search
It could be useful to have a tag system where you could label entries, or they could be auto-tagged when updating filters. This would allow you to tag entries by a project, or select entries for some kind of bulk action like update, export to bibtex, or delete.
You might also build a scoring system, e.g. for like/dislike tags.
litdb tag doi -t "tag1" "tag2" # add tag
litdb tab doi -r "tag" "tag2" # rm tagsThis would use your microphone to record and transcribe a query for search.
Do the search from the results. I did this with tesseract (https://pypi.org/project/pytesseract/)
import pyautogui
# Prompt the user to move the mouse to the first corner and press Enter
input("Move the mouse to the first corner and press Enter...")
x1, y1 = pyautogui.position()
# Prompt the user to move the mouse to the opposite corner and press Enter
input("Move the mouse to the opposite corner and press Enter...")
x2, y2 = pyautogui.position()
# Calculate the region
left = min(x1, x2)
top = min(y1, y2)
width = abs(x2 - x1)
height = abs(y2 - y1)
region = (left, top, width, height)
print(f"Selected region: {region}")
import pyscreeze
im = pyscreeze.screenshot(region=(left, top, width, height))
im.save('screenshot.png')
see mss also.
from PIL import Image
import pytesseract
# Open an image file
img = Image.open('screenshot.png')
# Use Tesseract to extract text
text = pytesseract.image_to_string(img)
# Print the extracted text
print(text)
This might be nice later when we have image embeddings.
litdb review --since '1 week ago'You need to have a way to review what comes in to litdb; it is part of learning about what is current. I currently do this with Emacs and scimax-org-feed. You could integrate review with update-filters, or by entries added in the past few days, or some other kind of query. Then you just need to add some format information to get what you want, e.g. org, maybe html?
select source, date_added from sources where date(date_added) > '2024-11-28' limit 5
litdb uses cosine similarity as the distance metric for the nearest neighbors. It might be useful to re-rank these with cross-encoding.
https://www.sbert.net/examples/applications/cross-encoder/README.html
- LitSuggest
- https://www.ncbi.nlm.nih.gov/research/litsuggest/
- Browser tool that suggests literature for you based on positive and negative PMIDs. Hosted by NIH.
- paper-qa
- https://github.com/Future-House/paper-qa
- This project by Andrew White uses LLM+RAG to explore a paper.
- ColBERT
- https://github.com/stanford-futuredata/ColBERT
- ColBERT is a fast retrieval model for large text collections. In theory it can probably be integrated with litdb. litdb is so simple, and works well enough so far without it.
Many of these projects require you to make an account. There are freemium levels in each one.
- ResearchRabbit
- https://www.researchrabbit.ai/
- This is a browser tool to navigate the scientific literature graphically. You can make collections, and papers that are related by citations are shown in a graph
- LitMaps
- https://www.litmaps.com/
- Another browser tool to graphically interact with scientific literature
- Keenious
- https://keenious.com/explore
- Browser / Google Docs and Word plugin. Finds related articles to the text in your document. I like Keenious when in Google Docs.
- scite.ai
- https://scite.ai/
- Browser tool that integrates GPT with the scientific literature, integration with Zotero
- Scopus AI
- https://www.scopus.com/search/form.uri?display=basic#scopus-ai
- Sponsored by Elsevier
- Dimensions AI
- https://app.dimensions.ai/discover/publication
- Seems similar to Scopus AI
- khoj
- https://khoj.dev/
- This is a desktop app that can be totally local, or in the cloud. It can index your files, and then you can chat with them. There is a freemium level.
- AnythingLLM
- https://anythingllm.com/
- Another tool that runs LLMs locally, and says it can index your files so you can chat with them.
- gpt4all
- https://www.nomic.ai/gpt4all
- Another tool that runs LLMs locally, and says it can index your files so you can chat with them.
With all these options, why does litdb exist? There are a lot of answers to that. First, I wanted to make it. I learned a lot about vector search by doing it. Second, I wanted a free, extensible solution for literature search that could also work for my local files while never putting data in the cloud, and that would work in Emacs. The projects above are very nice, easy to use, no or low-code solutions, and if that is what you are looking for, look there! If you want to hack on things yourself, look here.