Recent advances in remote sensing products allow near-real time monitoring of the Earth's surface. Despite increasing availability of near-daily time-series of satellite imagery, there has been little exploration of deep learning methods to utilize the unprecedented temporal density of observations. This may be particularly interesting in crop monitoring where time-series remote sensing data has been used frequently to exploit phenological differences of crops in the growing cycle over time. Therefore, we introduce DENETHOR: The DynamicEarthNET dataset for Harmonized, inter-Operabel, analysis-Ready, daily crop monitoring from space. Our dataset contains daily, analysis-ready Planet Fusion data together with Sentinel- 1 and 2 time-series for crop type classification in Brandenburg, Germany. Our baseline experiments underline that incorporating the available spatial and temporal information fully may not be straightforward and could require the design of tailored architectures. The dataset presents two main challenges to the community: Exploit the temporal dimension for improved crop classification and ensure that models can handle a domain shift to a different year.
If you use this dataset, please cite our paper:
@article{kondmann2021denethor,
title={DENETHOR: The DynamicEarthNET dataset for Harmonized, inter-Operable, analysis-Ready, daily crop monitoring from space},
author={Kondmann, Lukas and Toker, Aysim and Ru{\ss}wurm, Marc and Camero, Andr{\'e}s and Peressuti, Devis and Milcinski, Grega and Mathieu, Pierre-Philippe and Long{\'e}p{\'e}, Nicolas and Davis, Timothy and Marchisio, Giovanni and others},
journal = {NeurIPS Track on Datasets and Benchmarks},
year={2021}
}
We provide a combination of harmonized, declouded, daily Planet Fusion data at 3m resolution together with Sentinel-1 and 2 time series for high-quality field boundaries and crop ids in Brandenburg, Germany. Train and test tiles are spatially separated and taken from different years to encourage out-of-year generalization. Both tiles are identical in size with 24km × 24km.
Download our dataset as part of the AI4EO Food Security Challenge here.
The full dataset including the test data will be released after the challenge.
Update 09/22: Access the full challenge dataset including the test data via Radiant MLHub here. Due to a quota limit at Sync&Share, download of the ablation study data is temporarily disabled. I will update again once access is restored. This affects only the train 2019 and 2018 test data. The main dataset is unaffected by this.
Update 03/22: Access the full challenge dataset including the test data via Radiant MLHub here. If you're interested in the ablation study data (train tile year 2019 and test tile year 2018) you can find it here. The pw is dailycrops.
Also check out a similar dataset we're involved in building for South Africa here.
The field boundaries and crop type information is taken from the German state of Brandenburg. This data is collected as part of the Common Agricultural Policy of the European Union. Farmers self-report the crops they grow on their fields to receive subsidies. In Germany, this data is controlled and administered on the state level. The data is not only geograph- ically precise but also of high quality since a variety of checks via in-situ measurements or earth observation data can potentially expose cheating.
The raw crop information provides the fields in vector format with a crop id coded between 1-999. Fields with areas below 1km2 are excluded since they are often broken in shape and can not easily be incorporated. We aggregate the crop type into a limited set of high-level classes. The nine classes with training frequencies in brackets are: Wheat (305), Rye (276), Barley (137), Oats (45), Corn (251), Oil Seeds (201), Root Crops (23), Meadows (954) and Forage Crops (339).
Crops that do not fit into these categories are rare in the reference data but in these instances, we remove the respective fields instead of collecting them in a tenth ‘Other’ class. The class imbalance provides a challenge for machine learning algorithms but it is representative of the geographic region and an imbalance is generally common in real-world crop type mapping tasks.
The main source of imagery is the Fusion Monitoring product 2 by Planet Labs, a commercial provider of high-resolution satellite imagery. It is based on the Planetscope constellation of Cubesats which collect images of the Earth from over 180 small satellites. The product has a spatial resolution of 3m and collects 4 spectral bands (RGB + Near-infrared (NIR)). It has several features that make it promising for crop monitoring.
First, it provides imagery in a unique daily time interval which allows studying the evolution of crops in unprecedented temporal density. Second, the time series is an analysis-ready data (ARD), level 3 product. It delivers a temporally consistent collection of images with removed clouds and shadows. Third, it is a Harmonized Landsat Sentinel-2 (HLS) time series, meaning that the data is interoperable with Landsat and Sentinel-2 data.
To combine and compare Planet and the publicly available Sentinel data, we include imagery from Sentinel-1 and Sentinel-2 to the train and test tile. While the spatial and temporal depth of S2 is comparably low, the combination of spectral depth (S2) with spatial and temporal depth (Planet Fusion) may provide additional opportunities for crop type mapping.
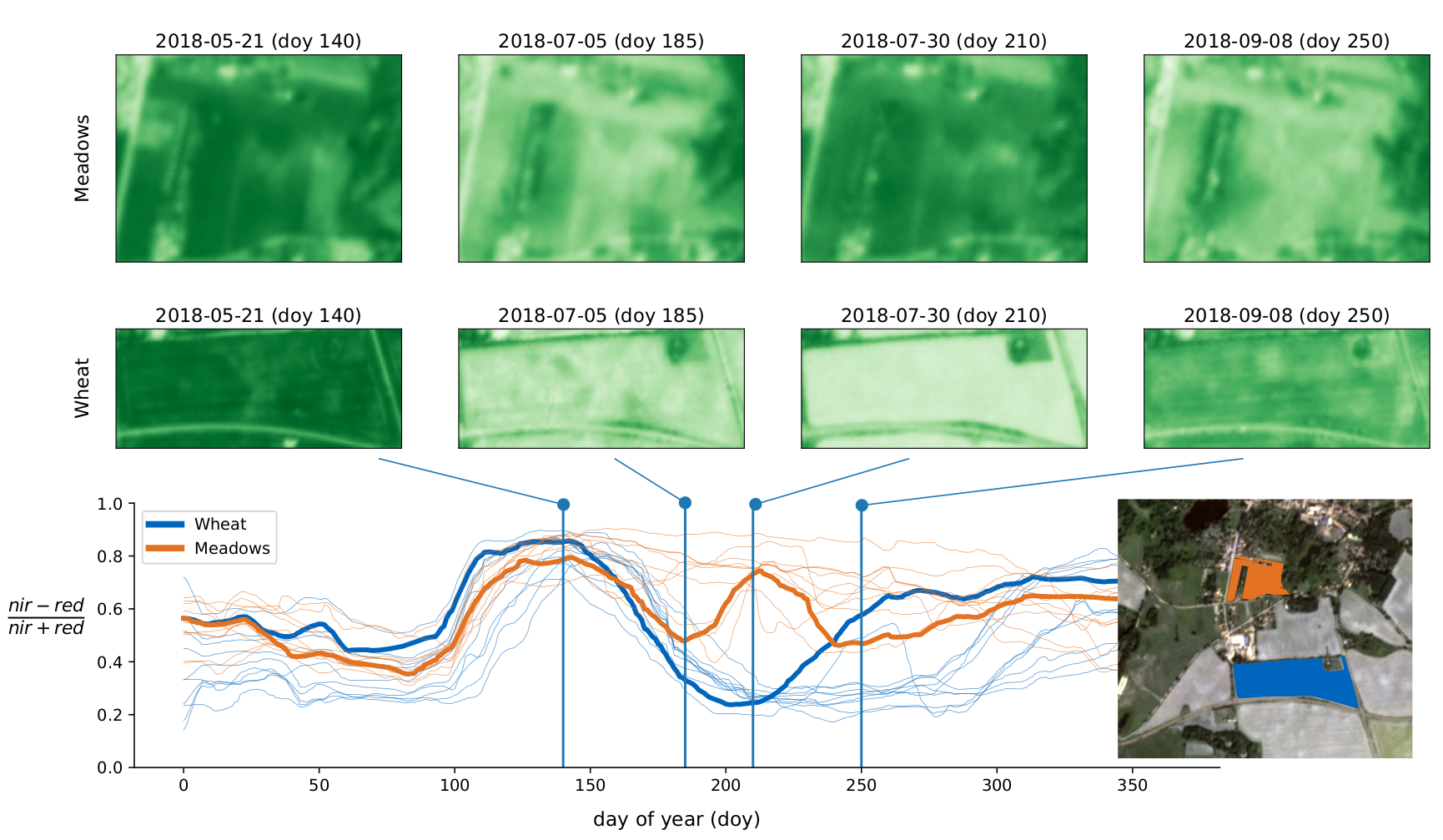 Examples of meadow and wheat parcels from the training dataset. The upper images show
two parcels at four (of 365) acquisition times. The bottom plot shows the Normalized Difference
Vegetation Index NDVI averaged over all pixels of these parcels. For reference, other fields
meadow and wheat fields are plotted in thin lines which illustrates that wheat and meadow vary
systematically between day 180 and 240. Meadows remain photo-synthetically active (high NDVI)
while wheat has been harvested.
Examples of meadow and wheat parcels from the training dataset. The upper images show
two parcels at four (of 365) acquisition times. The bottom plot shows the Normalized Difference
Vegetation Index NDVI averaged over all pixels of these parcels. For reference, other fields
meadow and wheat fields are plotted in thin lines which illustrates that wheat and meadow vary
systematically between day 180 and 240. Meadows remain photo-synthetically active (high NDVI)
while wheat has been harvested.
pip install -U -r requirements.txt
python train.py
mobilenet_v3_small #Spatial Encoder. If "none" => pixel average
transformermodel #Temporal Encoder
--logdir /tmp
--datapath /ssd/DENETHOR/PlanetL3H/Train/PF-SR
--labelgeojson /ssd/DENETHOR/crops_train_2018.geojson
--batchsize 12 --workers 8
requires a model checkpoint in the log directory
python test.py
mobilenet_v3_small #Spatial Encoder. If "none" => pixel average
transformermodel #Temporal Encoder
--logdir /tmp
--datapath /ssd/DENETHOR/PlanetL3H/Test/PF-SR
--labelgeojson /ssd/DENETHOR/crops_test_2019.geojson
--batchsize 24
separate script for testing & training PSETAE models
python test_psetae.py
--modelname psetae
--logdir /tmp
--datapath /ssd/DENETHOR/PlanetL3H/Test/PF-SR
--labelgeojson /ssd/DENETHOR/crops_test_2019.geojson
--batchsize 24
In case you want to run this, adjust the respective paths in random_forest.py
test all spatio-temporal model combinations
pytest tests.py
test single configuration (e.g. resnet18 + lstm)
pytest tests.py -k "test_spatiotemporalmodel[resnet18-lstm]"
